5W4Y
 
 | | Crystal Structure of Riboflavin Lyase (RcaE) with cofactor FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Riboflavin Lyase | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
5W4Z
 
 | | Crystal Structure of Riboflavin Lyase (RcaE) with modified FMN and substrate Riboflavin | | Descriptor: | 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-3-O-phosphono-D-ribitol, RIBOFLAVIN, Riboflavin Lyase | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
5W48
 
 | | Crystal Structure of Riboflavin Lyase (RcaE) | | Descriptor: | Riboflavin Lyase, SULFATE ION | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
4HW4
 
 | |
4X0W
 
 | | The crystal structure of mupain-1-17 in complex with murinised human uPA | | Descriptor: | SULFATE ION, Urokinase-type plasminogen activator, mupain-1-17, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-24 | | Release date: | 2015-10-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
4X1P
 
 | | The crystal structure of mupain-1-17 in complex with murinised human uPA at pH4.6 | | Descriptor: | MUPAIN-1-17, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-10-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
6ASL
 
 | |
6ASK
 
 | |
6ATV
 
 | | The molecular mechanisms by which NS1 of the 1918 Spanish influenza A virus hijack host protein-protein interactions | | Descriptor: | Adapter molecule crk, proline-rich motif in IAV-NS1 | | Authors: | Shen, Q, Zeng, D, Zhao, B, Li, P, Cho, J.H. | | Deposit date: | 2017-08-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Molecular Mechanisms of Tight Binding through Fuzzy Interactions.
Biophys. J., 114, 2018
|
|
5KH8
 
 | |
1HOS
 
 | |
6V43
 
 | | Crystal structure of the flavin oxygenase with cofactor and substrate bound involved in folate catabolism | | Descriptor: | FAD/FMN-containing dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, pteridine-2,4(1H,3H)-dione | | Authors: | Begley, T.P, Adak, S, Zhao, B, Li, P. | | Deposit date: | 2019-11-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A novel flavoenzyme catalyzed Baeyer-Villiger type rearrangement in bacterial folic acid catabolic pathway
To Be Published
|
|
1HPS
 
 | | RATIONAL DESIGN, SYNTHESIS AND CRYSTALLOGRAPHIC ANALYSIS OF A HYDROXYETHYLENE-BASED HIV-1 PROTEASE INHIBITOR CONTAINING A HETEROCYCLIC P1'-P2' AMIDE BOND ISOSTERE | | Descriptor: | 2-[(1R,3S,4S)-1-BENZYL-4-[N-(BENZYLOXYCARBONYL)-L-VALYL]AMINO-3-PHENYLPENTYL]-4(5)-(2-METHYLPROPIONYL)IMIDAZOLE, HIV-1 PROTEASE | | Authors: | Abdel-Meguid, S, Zhao, B. | | Deposit date: | 1994-05-24 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design, synthesis, and crystallographic analysis of a hydroxyethylene-based HIV-1 protease inhibitor containing a heterocyclic P1'--P2' amide bond isostere.
J.Med.Chem., 37, 1994
|
|
6XJD
 
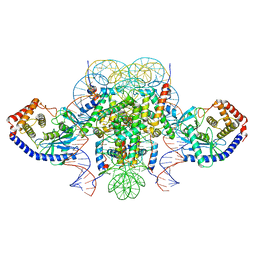 | | Two mouse cGAS catalytic domain binding to human assembled nucleosome | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Xu, P, Li, P, Zhao, B. | | Deposit date: | 2020-06-23 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
6U28
 
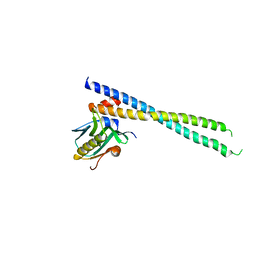 | | Crystal structure of 1918 NS1-ED W187A in complex with the p85-beta-iSH2 domain of human PI3K | | Descriptor: | Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Cho, J.H, Zhao, B, Savage, N, Li, P. | | Deposit date: | 2019-08-19 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UFX
 
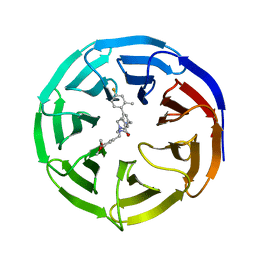 | | WD repeat-containing protein 5 complexed with N-[(3,5-dimethoxyphenyl)methyl]-4'-fluoro-5-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-2'-methyl[1,1'-biphenyl]-3-carboxamide (compound 13) | | Descriptor: | N-[(3,5-dimethoxyphenyl)methyl]-4'-fluoro-5-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-2'-methyl[1,1'-biphenyl]-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Rietz, T.A, Fesik, S.W, Zhao, B. | | Deposit date: | 2019-09-25 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Discovery and Structure-Based Optimization of Potent and Selective WD Repeat Domain 5 (WDR5) Inhibitors Containing a Dihydroisoquinolinone Bicyclic Core.
J.Med.Chem., 63, 2020
|
|
5FFO
 
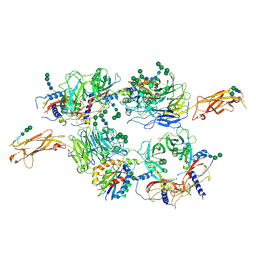 | | Integrin alpha V beta 6 in complex with pro-TGF-beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Zhao, B, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
1SBG
 
 | |
8TL8
 
 | | Structure of Orthoreovirus RNA Chaperone SigmaNS R6A mutant in complex with bile acid | | Descriptor: | GLYCOCHOLIC ACID, Protein sigma-NS | | Authors: | Prasad, B.V.V, Zhao, B, Hu, L, Neetu, N. | | Deposit date: | 2023-07-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of orthoreovirus RNA chaperone sigma NS, a component of viral replication factories.
Nat Commun, 15, 2024
|
|
8TL1
 
 | |
8TKA
 
 | |
1BGO
 
 | | CRYSTAL STRUCTURE OF CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PEPTIDOMIMETIC INHIBITOR | | Descriptor: | 1-[2-(3-BIPHENYL)-4-METHYLVALERYL)]AMINO-3-(2-PYRIDYLSULFONYL)AMINO-2-PROPANONE, CATHEPSIN K | | Authors: | Desjarlais, R.L, Yamashita, D.S, Oh, H.-J, Bondinell, W.E, Uzinskas, I.N, Erhard, K.F, Allen, A.C, Haltiwanger, R.C, Zhao, B, Smith, W.W, Abdel-Meguid, S.S, D'Alessio, K, Janson, C.A, Mcqueney, M.S, Tomaszek, T.A, Levy, M.A, Veber, D.F. | | Deposit date: | 1998-05-29 | | Release date: | 1999-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Use of X-Ray Co-Crystal Structures and Molecular Modeling to Design Potent and Selective Non-Peptide Inhibitors of Cathepsin K
J.Am.Chem.Soc., 120, 1998
|
|
5VPM
 
 | |
5VRP
 
 | |
5V8V
 
 | | Crystal Structure of Human Renin in Complex with a biphenylpipderidinylcarbinol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, methyl [(4S)-4-(3'-ethyl-6-fluoro[1,1'-biphenyl]-2-yl)-4-hydroxy-4-{(3R)-1-[4-(methylamino)butanoyl]piperidin-3-yl}butyl]carbamate | | Authors: | Concha, N, Zhao, B. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of renin inhibitors containing a simple aspartate binding moiety that imparts reduced P450 inhibition.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
