3UOE
 
 | | The crystal structure of dehydrogenase from Sinorhizobium meliloti | | Descriptor: | Dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-16 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | The crystal structure of dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
3U5T
 
 | | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase from Sinorhizobium meliloti | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase from Sinorhizobium meliloti
To be Published
|
|
3UXY
 
 | | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase/reductase SDR | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides
To be Published
|
|
3V2H
 
 | | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti | | Descriptor: | D-beta-hydroxybutyrate dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
3V76
 
 | | The crystal structure of a flavoprotein from Sinorhizobium meliloti | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavoprotein | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The crystal structure of a flavoprotein from Sinorhizobium meliloti
TO BE PUBLISHED
|
|
3VA6
 
 | |
3UBO
 
 | | The crystal structure of adenosine kinase from Sinorhizobium meliloti | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, adenosine kinase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | The crystal structure of adenosine kinase from Sinorhizobium meliloti
To be Published
|
|
3U9L
 
 | | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) from Sinorhizobium meliloti | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-19 | | Release date: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) from Sinorhizobium meliloti
TO BE PUBLISHED
|
|
3ZN3
 
 | | N-terminal domain of S. pombe Cdc23 APC subunit | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 8, MERCURY (II) ION | | Authors: | Zhang, Z, Yang, J, Conin, N, Kulkarni, K, Barford, D. | | Deposit date: | 2013-02-13 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Four Canonical Tpr Subunits of Human Apc/C Form Related Homo-Dimeric Structures and Stack in Parallel to Form a Tpr Suprahelix
J.Mol.Biol., 425, 2013
|
|
8HGO
 
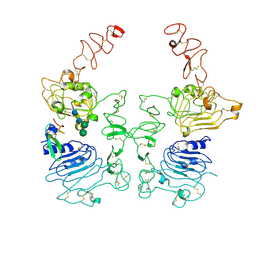 | | The EGF-bound EGFR/HER2 ectodomain complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor, ... | | Authors: | Zhang, Z, Bai, X. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
8HGP
 
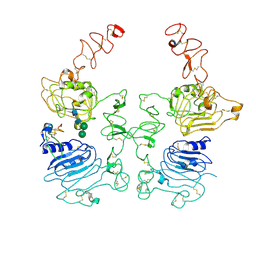 | | The EREG-bound EGFR/HER2 ectodomain complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Zhang, Z, Bai, X. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
8HGS
 
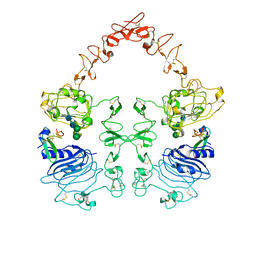 | | The EGF-bound EGFR ectodomain homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Pro-epidermal growth factor, ... | | Authors: | Zhang, Z, Bai, X. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
7YTP
 
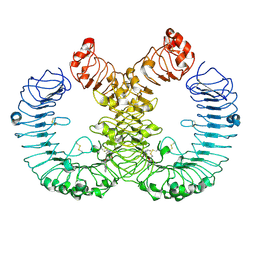 | | TLR7 in complex with an inhibitor | | Descriptor: | (2R,6R)-4-(8-cyanoquinolin-5-yl)-N-[(3S,4R)-4-fluoranylpyrrolidin-3-yl]-6-methyl-morpholine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2022-08-15 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | TLR7 in complex with an inhibitor
To Be Published
|
|
8HIJ
 
 | | The 5-MTHF-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HIK
 
 | | The TPP-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HII
 
 | | The BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | BRIL-SLC19A1 chimera, anti-BRIL Fab heavy chain, anti-BRIL Fab light chain, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
7E7X
 
 | |
8HD0
 
 | | Cell divisome sPG hydrolysis machinery FtsEX-EnvC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Zhang, Z, Chen, Y. | | Deposit date: | 2022-11-03 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural insight into the septal peptidoglycan hydrolysis machinery of bacterial cell division
To Be Published
|
|
8UX2
 
 | | Chromobacterium violaceum mono-ADP-ribosyltransferase CteC in complex with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, NAD(+)--protein-threonine ADP-ribosyltransferase, ... | | Authors: | Zhang, Z, Rondon, H, Das, C. | | Deposit date: | 2023-11-08 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of bacterial ubiquitin ADP-ribosyltransferase CteC reveals a substrate-recruiting insertion.
J.Biol.Chem., 300, 2023
|
|
7WGE
 
 | | Human NLRP1 complexed with thioredoxin | | Descriptor: | MAGNESIUM ION, NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2021-12-28 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for thioredoxin-mediated suppression of NLRP1 inflammasome.
Nature, 622, 2023
|
|
7DVA
 
 | | Structure of wild type Bt4394, a GH20 family sulfoglycosidase, in complex with 6S-GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DUP
 
 | | Apo structure of wild type Bt4394, a GH20 family sulfoglycosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVB
 
 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
8I96
 
 | | Cryo-EM structure of TLR7/TH-407b complex | | Descriptor: | (1S,3R)-5-[4-(8-nitroquinolin-5-yl)piperazin-1-yl]carbonyladamantan-2-one, Toll-like receptor 7 | | Authors: | Zhang, Z. | | Deposit date: | 2023-02-06 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of TLR7/TH-407b complex
To Be Published
|
|
7UZM
 
 | | Glutamate dehydrogenase 1 from human liver | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Zhang, Z. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
