5VF3
 
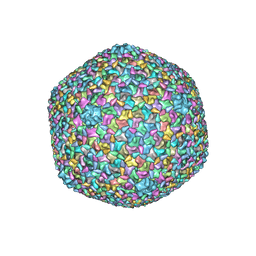 | | Bacteriophage T4 isometric capsid | | Descriptor: | Capsid vertex protein gp24, Highly immunogenic outer capsid protein, Major capsid protein, ... | | Authors: | Chen, Z, Sun, L, Zhang, Z, Fokine, A, Padilla-Sanchez, V, Hanein, D, Jiang, W, Rossmann, M.G, Rao, V.B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 isometric head at 3.3- angstrom resolution and its relevance to the assembly of icosahedral viruses.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8WX3
 
 | |
8WX2
 
 | |
8WX4
 
 | |
5X3G
 
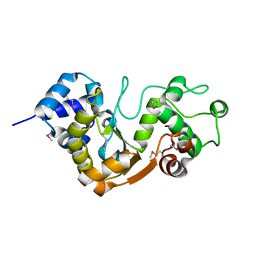 | |
7W7Y
 
 | | The crystal structure of human abl1 kinase domain in complex with ABL2-A5 | | Descriptor: | 5-[3-(5-methanoyl-2-methoxy-4-oxidanyl-phenyl)-1~{H}-pyrrolo[2,3-b]pyridin-5-yl]-~{N},~{N}-dimethyl-pyridine-3-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z.M. | | Deposit date: | 2021-12-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.20003033 Å) | | Cite: | Cell-Active, Reversible, and Irreversible Covalent Inhibitors That Selectively Target the Catalytic Lysine of BCR-ABL Kinase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
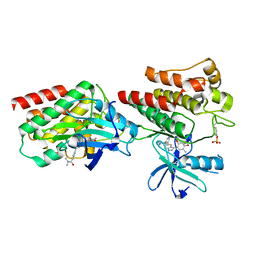
7W7X
 
 | | The crystal structure of human abl1 kinase domain in complex with ABL1-A11 | | Descriptor: | 5-[5-(dimethylcarbamoyl)pyridin-3-yl]-3-(5-fluorosulfonyloxy-2-methoxy-phenyl)-1H-pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z.M. | | Deposit date: | 2021-12-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.0000093 Å) | | Cite: | Cell-Active, Reversible, and Irreversible Covalent Inhibitors That Selectively Target the Catalytic Lysine of BCR-ABL Kinase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
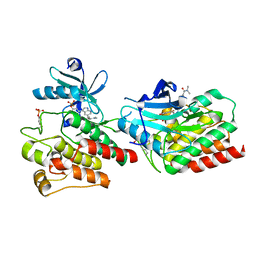
7X8R
 
 | | Cryo-EM structure of the Boc5-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-[(2-methylpropan-2-yl)oxycarbonylamino]phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X8S
 
 | | Cryo-EM structure of the WB4-24-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-(2-methylpropanoylamino)phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5X3H
 
 | |
8JIO
 
 | |
7KKH
 
 | | P1A4 Fab in complex with ARS1620 | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, P1A4 Fab Heavy Chain, P1A4 Fab Light Chain, ... | | Authors: | Basu, K, Rohweder, P.J, Zhang, Z, Bohn, M.-F, Shokat, K, Craik, C.S. | | Deposit date: | 2020-10-27 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A covalent inhibitor of K-Ras(G12C) induces MHC class I presentation of haptenated peptide neoepitopes targetable by immunotherapy.
Cancer Cell, 40, 2022
|
|
8I7T
 
 | | The crystal structure of human abl1 kinase domain in complex with ABL1-B4 | | Descriptor: | Tyrosine-protein kinase ABL1, [3-[5-[5-(dimethylcarbamoyl)pyridin-3-yl]-1H-pyrrolo[2,3-b]pyridin-3-yl]-4-methoxy-phenyl] ethanesulfonate | | Authors: | Zhu, C, Zhang, Z. | | Deposit date: | 2023-02-02 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80004954 Å) | | Cite: | Rationally designed BCR-ABL kinase inhibitors for improved leukemia treatment via covalent and pro-/dual-drug targeting strategies.
J Adv Res, 2024
|
|
8I7S
 
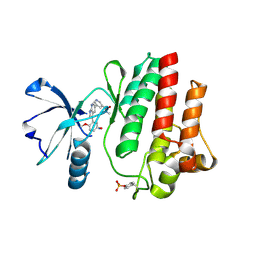 | | The crystal structure of human abl1 kinase domain in complex with ABL1-B1 | | Descriptor: | 5-[3-(2-methoxy-5-oxidanyl-phenyl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N,N-dimethyl-pyridine-3-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z. | | Deposit date: | 2023-02-02 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94635463 Å) | | Cite: | Rationally designed BCR-ABL kinase inhibitors for improved leukemia treatment via covalent and pro-/dual-drug targeting strategies.
J Adv Res, 2024
|
|
8I7Z
 
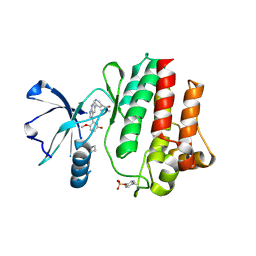 | | The crystal structure of human abl1 kinase domain in complex with ABL1-B5 | | Descriptor: | 5-[3-(2-methoxy-5-oxidanyl-phenyl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N,N-dimethyl-pyridine-3-carboxamide, BETA-ALANINE, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z. | | Deposit date: | 2023-02-02 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25495815 Å) | | Cite: | Rationally designed BCR-ABL kinase inhibitors for improved leukemia treatment via covalent and pro-/dual-drug targeting strategies.
J Adv Res, 2024
|
|
2ONC
 
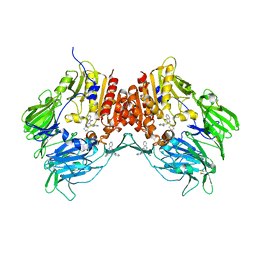 | | Crystal structure of human DPP-4 | | Descriptor: | 2-({2-[(3R)-3-AMINOPIPERIDIN-1-YL]-4-OXOQUINAZOLIN-3(4H)-YL}METHYL)BENZONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feng, J, Zhang, Z, Wallace, M.B, Stafford, J.A, Kaldor, S.W, Kassel, D.B, Navre, M, Shi, L, Skene, R.J, Asakawa, T, Takeuchi, K, Xu, R, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2007-01-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of alogliptin: a potent, selective, bioavailable, and efficacious inhibitor of dipeptidyl peptidase IV.
J.Med.Chem., 50, 2007
|
|
7W02
 
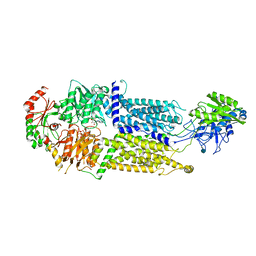 | | Cryo-EM structure of ATP-bound ABCA3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xie, T, Zhang, Z.K, Yue, J, Gong, X. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human surfactant lipid transporter ABCA3.
Sci Adv, 8, 2022
|
|
7W01
 
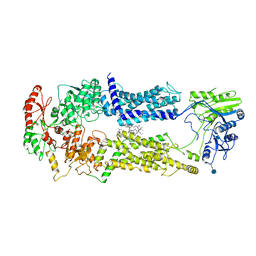 | | Cryo-EM structure of nucleotide-free ABCA3 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xie, T, Zhang, Z.K, Yue, J, Gong, X. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human surfactant lipid transporter ABCA3.
Sci Adv, 8, 2022
|
|
4HGA
 
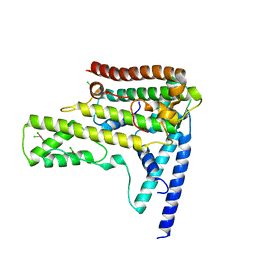 | | Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Liu, C.P, Xiong, C.Y, Wang, M.Z, Yu, Z.L, Yang, N, Chen, P, Zhang, Z.G, Li, G.H, Xu, R.M. | | Deposit date: | 2012-10-07 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7VA8
 
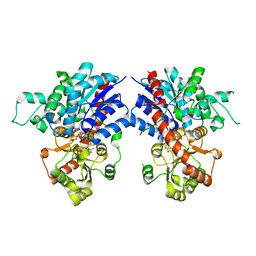 | | Crystal structure of MiCGT | | Descriptor: | UDP-glycosyltransferase 13, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Zhong, L, Zhang, Z.M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85003233 Å) | | Cite: | Directed Evolution of a Plant Glycosyltransferase for Chemo- and Regioselective Glycosylation of Pharmaceutically Significant Flavonoids
Acs Catalysis, 11, 2021
|
|
7VAA
 
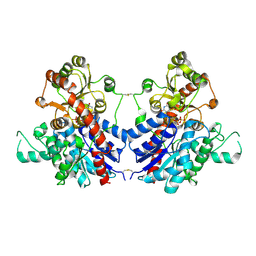 | |
7UW6
 
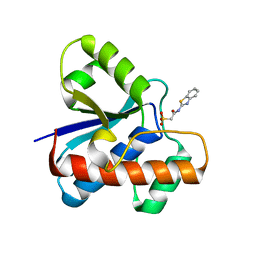 | | The co-crystal structure of low molecular weight protein tyrosine phosphatase (LMW-PTP) with a small molecule inhibitor SPAA-2 | | Descriptor: | 2-[(1,3-benzothiazol-2-yl)amino]-2-oxoethane-1-sulfonic acid, Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Wang, J, Zhang, Z.Y. | | Deposit date: | 2022-05-02 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Design of Active-Site-Directed, Highly Potent, Selective, and Orally Bioavailable Low-Molecular-Weight Protein Tyrosine Phosphatase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7C8U
 
 | | The crystal structure of COVID-19 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Luan, X, Shang, W, Wang, Y, Yin, W, Jiang, Y, Feng, S, Wang, Y, Liu, M, Zhou, R, Zhang, Z, Wang, F, Cheng, W, Gao, M, Wang, H, Wu, W, Tian, R, Tian, Z, Jin, Y, Jiang, H.W, Zhang, L, Xu, H.E, Zhang, S. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of COVID-19 main protease in complex with GC376
To Be Published
|
|
7U5J
 
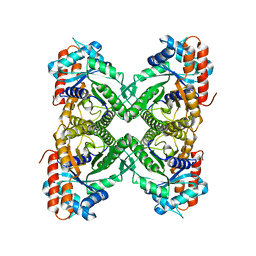 | | Cryo-EM Structure of ALDOA | | Descriptor: | Fructose-bisphosphate aldolase | | Authors: | Morgan, C.E, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
7U5M
 
 | | Cryo-EM Structure of GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Morgan, C.E, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
