6XRQ
 
 | | Structural descriptions of ligand interactions to DNA and RNA quadruplexes folded from the non-coding region of Pseudorabies virus | | 分子名称: | 2,7-bis[3-(morpholin-4-yl)propyl]-4,9-bis{[3-(morpholin-4-yl)propyl]amino}benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, POTASSIUM ION, RNA (5' GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A-3') | | 著者 | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | 登録日 | 2020-07-13 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Structural descriptions of ligand interactions to RNA quadruplexes folded from the non-coding region of Pseudorabies virus.
Biochimie, 2024
|
|
5X87
 
 | |
4GKU
 
 | |
4F83
 
 | | Crystal structure of the receptor binding domain of botulinum neurotoxin mosaic serotype C/D with a tetraethylene glycol molecule bound on the Hcn sub-domain and a sulfate ion at the putative active site | | 分子名称: | GLYCEROL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Zhang, Y, Buchko, G.W, Gardberg, A, Edwards, T.E, Sankaran, B, Robinson, H, Varnum, S.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2012-05-16 | | 公開日 | 2012-06-20 | | 最終更新日 | 2013-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insights into the functional role of the Hcn sub-domain of the receptor-binding domain of the botulinum neurotoxin mosaic serotype C/D.
Biochimie, 95, 2013
|
|
4H6P
 
 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a R101A substitution. | | 分子名称: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | 著者 | Zhang, Y, Robinson, H, Buchko, G.W. | | 登録日 | 2012-09-19 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.556 Å) | | 主引用文献 | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
6N33
 
 | | Crystal structure of fms kinase domain with a small molecular inhibitor, PLX5622 | | 分子名称: | 6-fluoro-N-[(5-fluoro-2-methoxypyridin-3-yl)methyl]-5-[(5-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]pyridin-2-amine, Macrophage colony-stimulating factor 1 receptor | | 著者 | Zhang, Y. | | 登録日 | 2018-11-14 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer's disease model.
Nat Commun, 10, 2019
|
|
4HS4
 
 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a Y129N substitution. | | 分子名称: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | 著者 | Zhang, Y, Robinson, H, Buchko, G.W. | | 登録日 | 2012-10-29 | | 公開日 | 2012-12-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
4NPU
 
 | |
5E17
 
 | |
5E18
 
 | |
4NPT
 
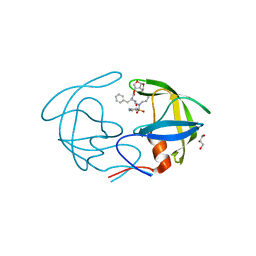 | | Crystal Structure of HIV-1 Protease Multiple Mutant P51 Complexed with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, GLYCEROL, Protease | | 著者 | Zhang, Y, Weber, I.T. | | 登録日 | 2013-11-22 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structures of darunavir-resistant HIV-1 protease mutant reveal atypical binding of darunavir to wide open flaps.
Acs Chem.Biol., 9, 2014
|
|
4OXW
 
 | |
9ATN
 
 | |
6XNZ
 
 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Target Capture Complex | | 分子名称: | 12RSS integration strand (34-mer), 12RSS non-integration strand (34-mer), 23RSS integration strand (45-mer), ... | | 著者 | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | 登録日 | 2020-07-05 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6XNY
 
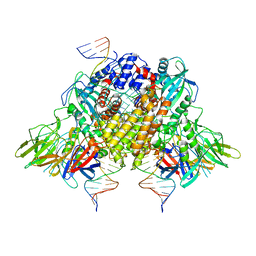 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Paired-Form) | | 分子名称: | 12RSS integration strand (55-mer), 12RSS signal DNA top strand (34-mer), 23RSS integration strand (66-mer), ... | | 著者 | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | 登録日 | 2020-07-05 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6Y5B
 
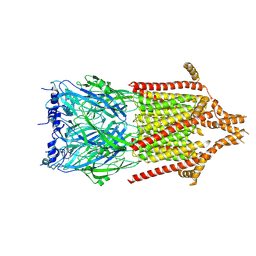 | | 5-HT3A receptor in Salipro (apo, asymmetric) | | 分子名称: | 5-hydroxytryptamine receptor 3A | | 著者 | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | 登録日 | 2020-02-25 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
6XNX
 
 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Dynamic-Form) | | 分子名称: | 12RSS integration strand DNA (55-MER), 12RSS signal top strand DNA (34-MER), 23RSS integration strand DNA (66-MER), ... | | 著者 | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | 登録日 | 2020-07-05 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6Y59
 
 | | 5-HT3A receptor in Salipro (apo, C5 symmetric) | | 分子名称: | 5-hydroxytryptamine receptor 3A | | 著者 | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | 登録日 | 2020-02-25 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
6Y5A
 
 | | Serotonin-bound 5-HT3A receptor in Salipro | | 分子名称: | 5-hydroxytryptamine receptor 3A, SEROTONIN | | 著者 | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | 登録日 | 2020-02-25 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
5WAN
 
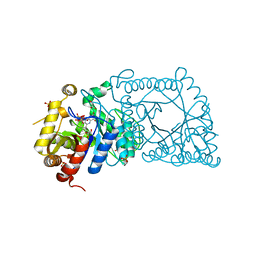 | | Crystal Structure of a flavoenzyme RutA in the pyrimidine catabolic pathway | | 分子名称: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Pyrimidine monooxygenase RutA, ... | | 著者 | Zhang, Y, Mukherjee, T, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | 登録日 | 2017-06-26 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.798 Å) | | 主引用文献 | Catalysis of a flavoenzyme-mediated amide hydrolysis.
J. Am. Chem. Soc., 132, 2010
|
|
5X21
 
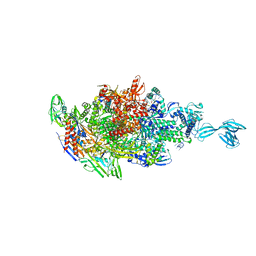 | | Crystal structure of Thermus thermophilus transcription initiation complex with GpA and pseudouridimycin (PUM) | | 分子名称: | (1S)-1,4-anhydro-5-[(N-carbamimidoylglycyl-N~2~-hydroxy-L-glutaminyl)amino]-5-deoxy-1-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-D-ribitol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Zhang, Y, Ebright, R. | | 登録日 | 2017-01-29 | | 公開日 | 2017-07-05 | | 最終更新日 | 2022-10-12 | | 実験手法 | X-RAY DIFFRACTION (3.323 Å) | | 主引用文献 | Antibacterial Nucleoside-Analog Inhibitor of Bacterial RNA Polymerase.
Cell, 169, 2017
|
|
6O7G
 
 | |
5X22
 
 | |
6I5X
 
 | | Crystal structure of Aspergillus fumigatus phosphomannomutase | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | 著者 | Zhang, Y, Raimi, O.G, Ferenbach, A.T, van Aalten, D.M.F. | | 登録日 | 2018-11-15 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of Aspergillus fumigatus phosphomannomutase
To Be Published
|
|
5UQ8
 
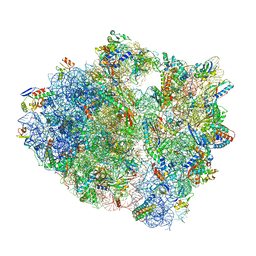 | | 70S ribosome complex with dnaX mRNA stem-loop and E-site tRNA ("out" conformation) | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | 登録日 | 2017-02-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2018-03-21 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
