7MOA
 
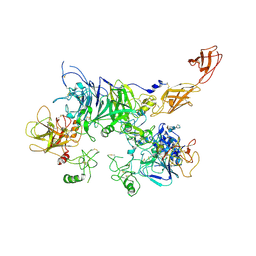 | | Cryo-EM structure of the c-MET II/HGF I complex bound with HGF II in a rigid conformation | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO7
 
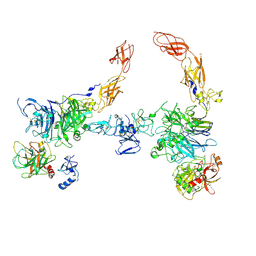 | | Cryo-EM structure of 2:2 c-MET/HGF holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO9
 
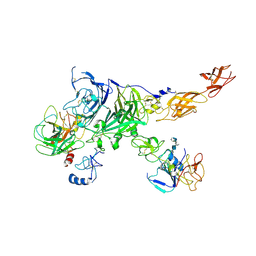 | | Cryo-EM map of the c-MET II/HGF I/HGF II (K4 and SPH) sub-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MOB
 
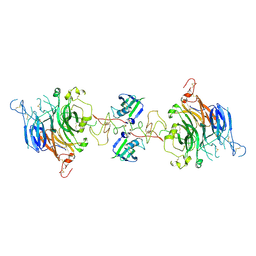 | | Cryo-EM structure of 2:2 c-MET/NK1 complex | | Descriptor: | Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO8
 
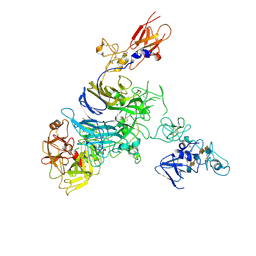 | | Cryo-EM structure of 1:1 c-MET I/HGF I complex after focused 3D refinement of holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
6Q2R
 
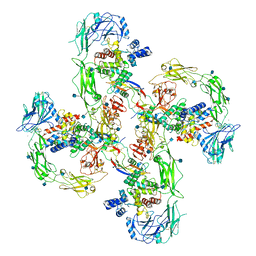 | | Cryo-EM structure of RET/GFRa2/NRTN extracellular complex in the tetrameric form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-2, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6Q2J
 
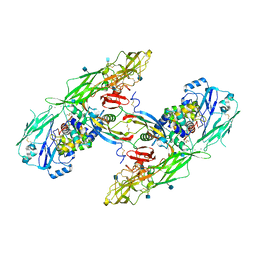 | | Cryo-EM structure of extracellular dimeric complex of RET/GFRAL/GDF15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-like, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6Q2S
 
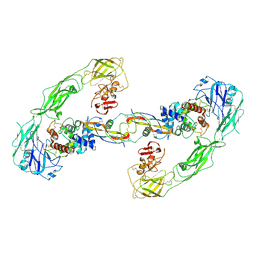 | | Cryo-EM structure of RET/GFRa3/ARTN extracellular complex. The 3D refinement was applied with C2 symmetry. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-3, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6Q2O
 
 | | Cryo-EM structure of RET/GFRa2/NRTN extracellular complex. The 3D refinement was applied with C2 symmetry. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-2, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6Q2N
 
 | | Cryo-EM structure of RET/GFRa1/GDNF extracellular complex | | Descriptor: | CALCIUM ION, GDNF family receptor alpha-1, Glial cell line-derived neurotrophic factor, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
4O67
 
 | | Human cyclic GMP-AMP synthase (cGAS) in complex with GAMP | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION, cGAMP | | Authors: | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O68
 
 | | Structure of human cyclic GMP-AMP synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O69
 
 | | Human cyclic GMP-AMP synthase (cGAS) in complex with sulfate ion | | Descriptor: | Cyclic GMP-AMP synthase, SULFATE ION, ZINC ION | | Authors: | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O6A
 
 | | Mouse cyclic GMP-AMP synthase (cGAS) in complex with DNA | | Descriptor: | Cyclic GMP-AMP synthase, DNA1, DNA2, ... | | Authors: | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4KSY
 
 | |
8SBD
 
 | | Cryo-EM structure of insulin amyloid-like fibril that is composed of two antiparallel protofilaments | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wang, L.W, Hall, C, Uchikawa, E, Chen, D.L, Choi, E, Zhang, X.W, Bai, X.C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of insulin fibrillation.
Sci Adv, 9, 2023
|
|
7XI6
 
 | | Crystal structure of C56 from pSSVi | | Descriptor: | C56 | | Authors: | Zhang, Z.F, Ren, Y, Chen, Y.Y, Zhang, X.W, Dong, Y.H, Gong, Y, Cao, P, Huang, L. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the AbrB-like protein C56 conserved in a novel family of integrated genetic elements in Sulfolobales
To Be Published
|
|
6W3I
 
 | | Crystal structure of a FAM46C mutant in complex with Plk4 | | Descriptor: | Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | Authors: | Chen, H, Shang, G.J, Lu, D.F, Zhang, X.W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
6W38
 
 | | Crystal structure of the FAM46C/Plk4 complex | | Descriptor: | Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | Authors: | Chen, H, Lu, D.F, Shang, G.J, Zhang, X.W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.48 Å) | | Cite: | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
6W3J
 
 | | Crystal structure of the FAM46C/Plk4/Cep192 complex | | Descriptor: | Centrosomal protein of 192 kDa, Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | Authors: | Chen, H, Lu, D.F, Shang, G.J, Zhang, X.W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.385 Å) | | Cite: | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
6W36
 
 | | Crystal structure of FAM46C | | Descriptor: | SULFATE ION, Terminal nucleotidyltransferase 5C | | Authors: | Shang, G.J, Zhang, X.W, Chen, H, Lu, D.F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
7VQD
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl pyrophosphate and geranylgeranyl pyrophosphate. | | Descriptor: | Di-trans-poly-cis-decaprenylcistransferase, FARNESYL DIPHOSPHATE, NerylNeryl pyrophosphate, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQC
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with pyrophosphate | | Descriptor: | Di-trans-poly-cis-decaprenylcistransferase, PYROPHOSPHATE 2-, SULFATE ION | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQ9
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl thiopyrophosphate and isopentyl S-thiolodiphosphate | | Descriptor: | 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, Di-trans-poly-cis-decaprenylcistransferase, MAGNESIUM ION, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQB
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl pyrophosphate and dimethylallyl diphosphate | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, FARNESYL DIPHOSPHATE, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
