6LWR
 
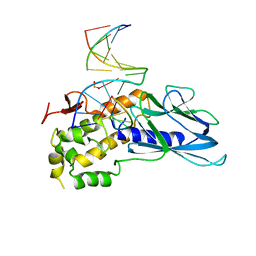 | | Crystal structure of human NEIL1(K242) bound to duplex DNA containing a cleaved C:T mismatch | | 分子名称: | DNA (5'-D(*CP*GP*TP*CP*CP*(PDA))-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | 著者 | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2020-02-07 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
5YC7
 
 | |
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (5.5 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6NT9
 
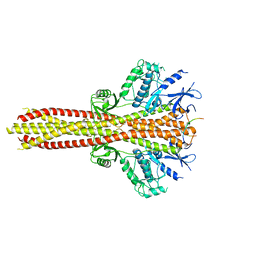 | | Cryo-EM structure of the complex between human TBK1 and chicken STING | | 分子名称: | Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | 著者 | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | 登録日 | 2019-01-28 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of STING binding with and phosphorylation by TBK1.
Nature, 567, 2019
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
5Z1C
 
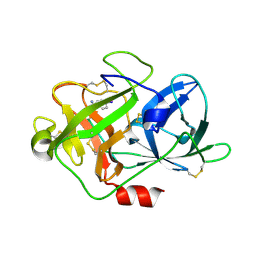 | | The crystal structure of uPA in complex with 4-Iodobenzylamine at pH7.4 | | 分子名称: | 1-(4-iodophenyl)methanamine, Urokinase-type plasminogen activator | | 著者 | Jiang, L.G, Zhang, X, Luo, Z.P, Huang, M.D. | | 登録日 | 2017-12-25 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 49, 2018
|
|
2KA9
 
 | | Solution structure of PSD-95 PDZ12 complexed with cypin peptide | | 分子名称: | Disks large homolog 4, cypin peptide | | 著者 | Wang, W.N, Weng, J.W, Zhang, X, Liu, M.L, Zhang, M.J. | | 登録日 | 2008-11-03 | | 公開日 | 2009-06-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Creating conformational entropy by increasing interdomain mobility in ligand binding regulation: a revisit to N-terminal tandem PDZ domains of PSD-95
J.Am.Chem.Soc., 131, 2009
|
|
8EW2
 
 | |
8EW0
 
 | |
6LJU
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-[[3-chloranyl-4-(methylamino)-2-phenyl-phenyl]amino]benzoic acid, Fatty acid-binding protein, ... | | 著者 | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | 登録日 | 2019-12-17 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJX
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | 分子名称: | 2-phenylazanylbenzoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | 登録日 | 2019-12-17 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJW
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-phenylazanylbenzoic acid, Fatty acid-binding protein, ... | | 著者 | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | 登録日 | 2019-12-17 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJT
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-[(3-chloranyl-2-phenyl-phenyl)amino]benzoic acid, Fatty acid-binding protein, ... | | 著者 | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | 登録日 | 2019-12-17 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
8SBD
 
 | | Cryo-EM structure of insulin amyloid-like fibril that is composed of two antiparallel protofilaments | | 分子名称: | Insulin A chain, Insulin B chain | | 著者 | Wang, L.W, Hall, C, Uchikawa, E, Chen, D.L, Choi, E, Zhang, X.W, Bai, X.C. | | 登録日 | 2023-04-03 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of insulin fibrillation.
Sci Adv, 9, 2023
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | 分子名称: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | 著者 | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2015-11-25 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
3F8I
 
 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group P21 | | 分子名称: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1 | | 著者 | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | 登録日 | 2008-11-12 | | 公開日 | 2009-01-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
6IYA
 
 | |
3F8J
 
 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group C222(1) | | 分子名称: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1, ... | | 著者 | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | 登録日 | 2008-11-12 | | 公開日 | 2009-01-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
6W36
 
 | | Crystal structure of FAM46C | | 分子名称: | SULFATE ION, Terminal nucleotidyltransferase 5C | | 著者 | Shang, G.J, Zhang, X.W, Chen, H, Lu, D.F. | | 登録日 | 2020-03-09 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.854 Å) | | 主引用文献 | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
6NT5
 
 | | Cryo-EM structure of full-length human STING in the apo state | | 分子名称: | Stimulator of interferon protein | | 著者 | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | 登録日 | 2019-01-28 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6W3I
 
 | | Crystal structure of a FAM46C mutant in complex with Plk4 | | 分子名称: | Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | 著者 | Chen, H, Shang, G.J, Lu, D.F, Zhang, X.W. | | 登録日 | 2020-03-09 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.802 Å) | | 主引用文献 | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
6W3J
 
 | | Crystal structure of the FAM46C/Plk4/Cep192 complex | | 分子名称: | Centrosomal protein of 192 kDa, Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | 著者 | Chen, H, Lu, D.F, Shang, G.J, Zhang, X.W. | | 登録日 | 2020-03-09 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (4.385 Å) | | 主引用文献 | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
6NT6
 
 | | Cryo-EM structure of full-length chicken STING in the apo state | | 分子名称: | Stimulator of interferon genes protein | | 著者 | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | 登録日 | 2019-01-28 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
