2KCU
 
 | | NMR solution structure of an uncharacterized protein from Chlorobium tepidum. Northeast Structural Genomics target CtR107 | | Descriptor: | protein CtR107 | | Authors: | Mills, J.L, Zhang, Q, Sukumaran, D.K, Wang, D, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of an uncharacterized protein from Chlorobium tepidum. Northeast Structural Genomics target CtR107
To be Published
|
|
2KTB
 
 | |
2KWO
 
 | | Solution structure of the double PHD (plant homeodomain) fingers of human transcriptional protein DPF3b bound to a histone H4 peptide containing N-terminal acetylation at Serine 1 | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-14 | | Release date: | 2010-07-14 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
2KWN
 
 | | Solution structure of the double PHD (plant homeodomain) fingers of human transcriptional protein DPF3b bound to a histone H4 peptide containing acetylation at Lysine 16 | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-13 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
2KWK
 
 | | Solution structures of the double PHD fingers of human transcriptional protein DPF3b bound to a H3 peptide wild type | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
3CXR
 
 | | Crystal structure of gluconate 5-dehydrogase from streptococcus suis type 2 | | Descriptor: | Dehydrogenase with different specificities | | Authors: | Peng, H, Gao, F, Zhang, Q, Liu, Y, Gao, G.F. | | Deposit date: | 2008-04-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the catalytic mechanism of gluconate 5-dehydrogenase from Streptococcus suis: Crystal structures of the substrate-free and quaternary complex enzymes.
Protein Sci., 18, 2009
|
|
7YIX
 
 | | The Cryo-EM Structure of Human Tissue Nonspecific Alkaline Phosphatase and Single-Chain Fragment Variable (ScFv) Complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, ... | | Authors: | Yu, Y.T, Yao, D.Q, Zhang, Q, Rao, B, Xia, Y, Lu, Y, Qin, A, Ma, P.X, Cao, Y. | | Deposit date: | 2022-07-18 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | The structural pathology for hypophosphatasia caused by malfunctional tissue non-specific alkaline phosphatase.
Nat Commun, 14, 2023
|
|
3G60
 
 | | Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding | | Descriptor: | (4R,11R,18R)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1a | | Authors: | Aller, S.G, Yu, J, Ward, A, Weng, Y, Chittaboina, S, Zhuo, R, Harrell, P.M, Trinh, Y.T, Zhang, Q, Urbatsch, I.L, Chang, G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science, 323, 2009
|
|
3G5U
 
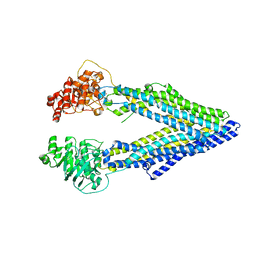 | | Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding | | Descriptor: | MERCURY (II) ION, Multidrug resistance protein 1a | | Authors: | Aller, S.G, Yu, J, Ward, A, Weng, Y, Chittaboina, S, Zhuo, R, Harrell, P.M, Trinh, Y.T, Zhang, Q, Urbatsch, I.L, Chang, G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science, 323, 2009
|
|
3G61
 
 | | Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding | | Descriptor: | (4S,11S,18S)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1a | | Authors: | Aller, S.G, Yu, J, Ward, A, Weng, Y, Chittaboina, S, Zhuo, R, Harrell, P.M, Trinh, Y.T, Zhang, Q, Urbatsch, I.L, Chang, G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science, 323, 2009
|
|
7YIV
 
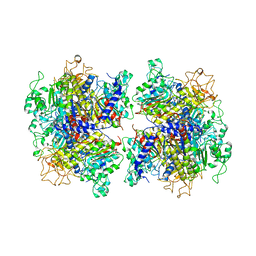 | | The Crystal Structure of Human Tissue Nonspecific Alkaline Phosphatase (ALPL) at Basic pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, ... | | Authors: | Cao, Y, Qin, A, Yu, Y.T, Yao, D.Q, Zhang, Q, Rao, B, Xia, Y, Lu, Y. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | The structural pathology for hypophosphatasia caused by malfunctional tissue non-specific alkaline phosphatase.
Nat Commun, 14, 2023
|
|
7YIW
 
 | | The Crystal Structure of Human Tissue Nonspecific Alkaline Phosphatase (ALPL) at Acidic pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, ... | | Authors: | Yu, Y.T, Yao, D.Q, Zhang, Q, Rao, B, Xia, Y, Lu, Y, Qin, A, Cao, Y. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The structural pathology for hypophosphatasia caused by malfunctional tissue non-specific alkaline phosphatase.
Nat Commun, 14, 2023
|
|
6O30
 
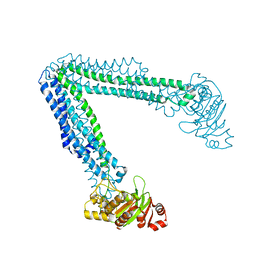 | | Lipid A transporter MsbA from Salmonella typhimurium | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Padayatti, P.S, Zhang, Q, Wilson, I.A, Lee, S.C, Stanfield, R.L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.47 Å) | | Cite: | Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 2019
|
|
8I8A
 
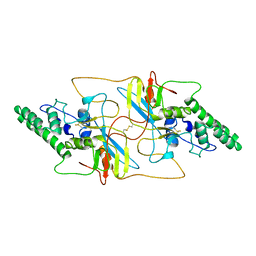 | |
8I8C
 
 | |
8I8B
 
 | | Outer shell and inner layer structures of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) | | Descriptor: | 38K, AcOrf-109 peptide, Early 49 Daa protein, ... | | Authors: | Jia, X, Gao, Y, Zhang, Q. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Architecture of the baculovirus nucleocapsid revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7VH8
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Zhao, Y, Zhang, Q, Yang, H, Rao, Z. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332.
Protein Cell, 13, 2022
|
|
5VN9
 
 | | Structure of bacteriorhodopsin from crystals grown at 4 deg C using GlyNCOC15+4 as an LCP host lipid | | Descriptor: | Bacteriorhodopsin | | Authors: | Ishchenko, A, Peng, L, Zinovev, E, Vlasov, A, Lee, S.C, Kuklin, A, Mishin, A, Borshchevskiy, V, Zhang, Q, Cherezov, V. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Chemically Stable Lipids for Membrane Protein Crystallization.
Cryst Growth Des, 17, 2017
|
|
5VN7
 
 | | Structure of bacteriorhodopsin from crystals grown at 20 deg Celcius using GlyNCOC15+4 as an LCP host lipid | | Descriptor: | Bacteriorhodopsin | | Authors: | Ishchenko, A, Peng, L, Zinovev, E, Vlasov, A, Lee, S.C, Kuklin, A, Mishin, A, Borshchevskiy, V, Zhang, Q, Cherezov, V. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chemically Stable Lipids for Membrane Protein Crystallization.
Cryst Growth Des, 17, 2017
|
|
7BVD
 
 | | Anthranilate synthase component I (TrpE)[Mycolicibacterium smegmatis] | | Descriptor: | Anthranilate synthase component 1, BENZOIC ACID, GLYCEROL, ... | | Authors: | Chen, Y, Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of subunit I of the anthranilate synthase complex of Mycolicibacterium smegmatis
Biochem.Biophys.Res.Commun., 527, 2020
|
|
7ZKA
 
 | | ABCB1 V978C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Urbatsch, I, Zhang, Q, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK9
 
 | | ABCB1 L971C mutant (mABCB1) in the inward facing state | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1 | | Authors: | Parey, K, Januliene, D, Gewering, T, Zhang, Q, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
5WCH
 
 | | Crystal structure of the catalytic domain of human USP9X | | Descriptor: | Probable ubiquitin carboxyl-terminal hydrolase FAF-X, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Zhang, Q, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and activity-based labeling reveal the mechanisms for linkage-specific substrate recognition by deubiquitinase USP9X.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5X0R
 
 | | Crystal Structure of PXR LBD Complexed with SJB7 | | Descriptor: | 4-[(4-tert-butylphenyl)sulfonyl]-1-(2,4-dimethoxy-5-methylphenyl)-5-methyl-1H-1,2,3-triazole, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Lv, L, Lin, W, Chai, S.C, Zhang, Q, Chen, T. | | Deposit date: | 2017-01-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.665 Å) | | Cite: | SPA70 is a potent antagonist of human pregnane X receptor.
Nat Commun, 8, 2017
|
|
3SYY
 
 | | Crystal Structure of an alkaline exonuclease (LHK-Exo) from Laribacter hongkongensis | | Descriptor: | Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
