5XME
 
 | | Solution structure of C-terminal domain of TRADD | | Descriptor: | Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Lin, Z, Zhang, N. | | Deposit date: | 2017-05-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain of TRADD reveals a novel fold in the death domain superfamily.
Sci Rep, 7, 2017
|
|
5XTA
 
 | | Crystal structure of lpg1832, a VirK family protein from Legionella pneumophila | | Descriptor: | GLYCEROL, SULFATE ION, VirK protein | | Authors: | Yin, S, Gong, X, Zhang, N, Ge, H. | | Deposit date: | 2017-06-18 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of lpg1832, a VirK family protein from Legionella pneumophila, reveals a novel fold for bacterial VirK proteins
FEBS Lett., 591, 2017
|
|
6SES
 
 | | Tubulin-B2 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Guo, B, Rodriguez-Gabin, A, Prota, A.E, Muehlethaler, T, Zhang, N, Ye, K, Steinmetz, M.O, Band Horwitz, S, Smith III, A.B, McDaid, H.M. | | Deposit date: | 2019-07-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Refinement of the Tubulin Ligand (+)-Discodermolide to Attenuate Chemotherapy-Mediated Senescence.
Mol.Pharmacol., 98, 2020
|
|
2ASS
 
 | | Crystal structure of the Skp1-Skp2-Cks1 complex | | Descriptor: | BENZAMIDINE, Cyclin-dependent kinases regulatory subunit 1, PHOSPHATE ION, ... | | Authors: | Hao, B, Zhang, N, Schulman, B.A, Wu, G, Pagano, M, Pavletich, N.P. | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of the Cks1-Dependent Recognition of p27(Kip1) by the SCF(Skp2) Ubiquitin Ligase.
Mol.Cell, 20, 2005
|
|
2AST
 
 | | Crystal structure of Skp1-Skp2-Cks1 in complex with a p27 peptide | | Descriptor: | BENZAMIDINE, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Hao, B, Zhang, N, Schulman, B.A, Wu, G, Pagano, M, Pavletich, N.P. | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Cks1-Dependent Recognition of p27(Kip1) by the SCF(Skp2) Ubiquitin Ligase.
Mol.Cell, 20, 2005
|
|
5H5Z
 
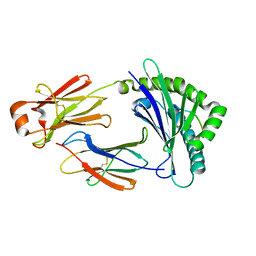 | | Crystal structure of bony fish MHC class I, peptide and B2m II | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide chain | | Authors: | Chen, Z, Zhang, N, Qi, J, Li, X, Chen, R, Wang, Z, Gao, F.G, Xia, C. | | Deposit date: | 2016-11-10 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Mechanism of beta 2m Molecule-Induced Changes in the Peptide Presentation Profile in a Bony Fish.
Iscience, 23, 2020
|
|
9IJU
 
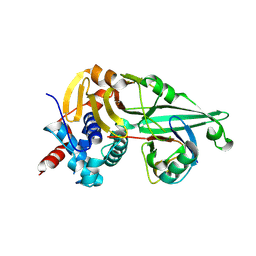 | | Sertraline enhances the deubiquitinase activity of USP7 by binding to its switching loop region | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Shi, L, Xu, Z, Chen, X, Xiong, B, Zhang, N. | | Deposit date: | 2024-06-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Sertraline and Astemizole Enhance the Deubiquitinase Activity of USP7 by Binding to Its Switching Loop Region.
J.Med.Chem., 68, 2025
|
|
9IML
 
 | | Sertraline enhances the deubiquitinase activity of USP7 by binding to its switching loop region | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Shi, L, Xu, Z, Chen, X, Xiong, B, Zhang, N. | | Deposit date: | 2024-07-03 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Sertraline and Astemizole Enhance the Deubiquitinase Activity of USP7 by Binding to Its Switching Loop Region.
J.Med.Chem., 68, 2025
|
|
5HKW
 
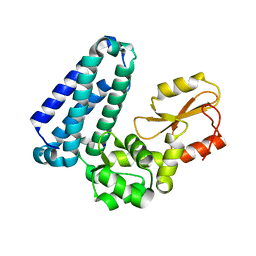 | | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution
To be published
|
|
5HKY
 
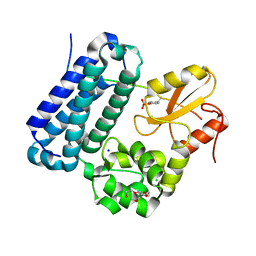 | | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase CBL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form)
To be published
|
|
5HKX
 
 | | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CBL, SODIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution
To be published
|
|
5HL0
 
 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION, Sprouty 2 (SPRY2) | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution
To Be Published
|
|
5HKZ
 
 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form) | | Descriptor: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form)
To be published
|
|
8HJE
 
 | | Vismodegib binds to the catalytical domain of human Ubiquitin-Specific Protease 28 | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Shi, L, Wang, H, Xu, Z, Xiong, B, Zhang, N. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based discovery of potent USP28 inhibitors derived from Vismodegib.
Eur.J.Med.Chem., 254, 2023
|
|
6KNM
 
 | | Apelin receptor in complex with single domain antibody | | Descriptor: | Apelin receptor,Rubredoxin,Apelin receptor, Single domain antibody JN241, ZINC ION | | Authors: | Ma, Y.B, Ding, Y, Song, X, Ma, X, Li, X, Zhang, N, Song, Y, Sun, Y, Shen, Y, Zhong, W, Hu, L.A, Ma, Y.L, Zhang, M.Y. | | Deposit date: | 2019-08-06 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-guided discovery of a single-domain antibody agonist against human apelin receptor.
Sci Adv, 6, 2020
|
|
8KB0
 
 | | Crystal structure of 01JD-AEAIIVAMV | | Descriptor: | Beta2-microglobulin, MHC class I antigen alpha chain, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
8KB1
 
 | | Crystal structure of 11JD | | Descriptor: | Beta2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
8WFM
 
 | | Crystal structure of Omicron BA.1 in complex with a neutralizing antibody scFv T11 | | Descriptor: | Spike protein S1, T11 scFv | | Authors: | Terekhov, S.S, Mokrushina, Y.A, Zhang, M, Zhang, N, Gabibov, A, Guo, Y. | | Deposit date: | 2023-09-19 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of Omicron BA.1 in complex with a neutralizing antibody scFv T11
To Be Published
|
|
8WFH
 
 | | Crystal structure of Omicron BA.4/5 in complex with a neutralizing antibody scFv D1 | | Descriptor: | D1 scFv, Spike protein S1 | | Authors: | Terekhov, S.S, Mokrushina, Y.A, Zhang, M, Zhang, N, Gabibov, A, Guo, Y. | | Deposit date: | 2023-09-19 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of Omicron BA.4/5 in complex with a neutralizing antibody scFv D1
To Be Published
|
|
5H94
 
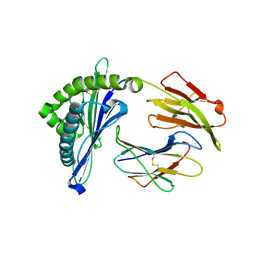 | | Crystal structure of Swine MHC CLASSI for 1.48 angstroms | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Nonapeptide from Influenza A virus HA protein | | Authors: | Fan, S, Zhang, N, Wang, S, Wu, Y, Xia, C. | | Deposit date: | 2015-12-25 | | Release date: | 2016-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Biochemical Analyses of Swine Major Histocompatibility Complex Class I Complexes and Prediction of the Epitope Map of Important Influenza A Virus Strains
J.Virol., 90, 2016
|
|
2MUU
 
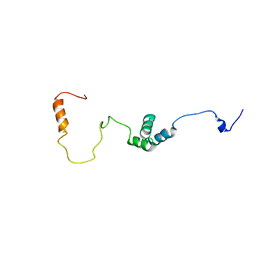 | |
2MUX
 
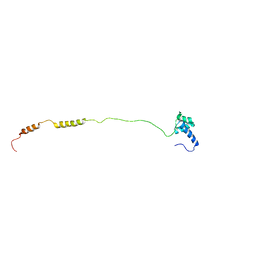 | |
8J4G
 
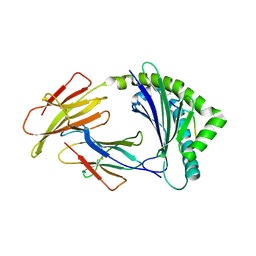 | | Crystal structure of 11JD mutant-I62N | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-04-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of duck MHC 11JD mutant-I62N
To Be Published
|
|
8JQT
 
 | | Crystal structure of U03-GPAKGIEYD | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-06-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of duck MHC U03
To Be Published
|
|
8KCV
 
 | | Crystal structure of UDA01-CAAMDDFQL | | Descriptor: | Beta2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of duck MHC UDA01
To Be Published
|
|
