8YFX
 
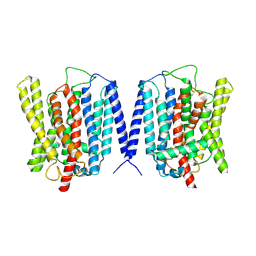 | | Cryo EM structure of human phosphate channel XPR1 at inward-facing state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 2024
|
|
8YFW
 
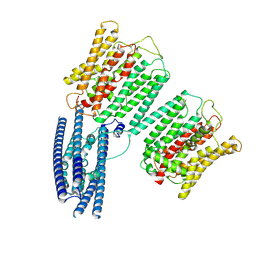 | | Cryo EM structure of human phosphate channel XPR1 at intermediate state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 2024
|
|
9IWS
 
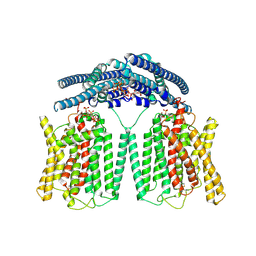 | | Cryo EM structure of human phosphate channel XPR1 in complex with IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 2024
|
|
4X2A
 
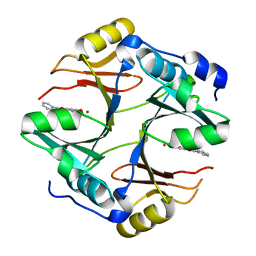 | | Crystal structure of mouse glyoxalase I complexed with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L, Li, C, Zhao, Y, Hu, X. | | Deposit date: | 2014-11-26 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In Vitro Inhibition of Glyoxalase І by Flavonoids: New Insights from Crystallographic Analysis.
Curr Top Med Chem, 16, 2016
|
|
3B8U
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
4RHD
 
 | | DNA Duplex with Novel ZP Base Pair | | Descriptor: | DNA 9mer novel P nucleobase, DNA 9mer novel Z nucleobase, MAGNESIUM ION | | Authors: | Zhang, W, Zhang, L, Benner, S, Huang, Z. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of functional six-nucleotide DNA.
J.Am.Chem.Soc., 137, 2015
|
|
3ZWM
 
 | | Crystal structure of ADP ribosyl cyclase complexed with substrate NAD and product cADPR | | Descriptor: | ADP-RIBOSYL CYLCASE, CYCLIC ADENOSINE DIPHOSPHATE-RIBOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWO
 
 | | Crystal structure of ADP ribosyl cyclase complexed with reaction intermediate | | Descriptor: | 3-(AMINOCARBONYL)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYD ROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2- YL]PYRIDINIUM, ADP-RIBOSYL CYCLASE, GUANOSINE DIPHOSPHATE RIBOSE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWY
 
 | | Crystal structure of ADP-ribosyl cyclase complexed with 8-bromo-ADP- ribose and cyclic 8-bromo-cyclic-ADP-ribose | | Descriptor: | (2R,3R,4S,5R,13R,14S,15R,16R)-24-amino-18-bromo-3,4,14,15-tetrahydroxy-7,9,11,25,26-pentaoxa-17,19,22-triaza-1-azonia-8 ,10-diphosphapentacyclo[18.3.1.1^2,5^.1^13,16^.0^17,21^]hexacosa-1(24),18,20,22-tetraene-8,10-diolate 8,10-dioxide, ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-8-bromo-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4S)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
To be Published
|
|
3ZWX
 
 | | Crystal structure of ADP-ribosyl cyclase complexed with 8-bromo-ADP- ribose | | Descriptor: | ADP-RIBOSYL CYCLASE, CHLORIDE ION, [(2R,3S,4R,5R)-5-(6-amino-8-bromo-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4S)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
To be Published
|
|
3ZWW
 
 | | Crystal structure of ADP-ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.3 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWV
 
 | | Crystal structure of ADP-ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.3 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of intermediates along the cyclization pathway of Aplysia ADP-ribosyl cyclase.
J. Mol. Biol., 415, 2012
|
|
7REF
 
 | | Structure of MS3494 from Mycobacterium smegmatis | | Descriptor: | BROMIDE ION, MS3494 | | Authors: | Kent, J.E, Aleshin, A.E, Zhang, L, Niederweis, M, Marassi, F.M. | | Deposit date: | 2021-07-12 | | Release date: | 2021-08-18 | | Last modified: | 2022-06-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A periplasmic cinched protein is required for siderophore secretion and virulence of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
3ZWP
 
 | | Crystal structure of ADP ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.1 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, GLYCEROL, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
7QFV
 
 | | Crystal structure of KLK6 in complex with compound 17a | | Descriptor: | KLK6 Activity-Based Probe (Ahx-DPhe-Ser(Z)-Dht-Arg-DPP), Kallikrein-6 | | Authors: | Jagtap, P.K.A, Zhang, L, De Vita, E, Tate, E.W, Hennig, J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A KLK6 Activity-Based Probe Reveals a Role for KLK6 Activity in Pancreatic Cancer Cell Invasion.
J.Am.Chem.Soc., 144, 2022
|
|
3JAU
 
 | | The cryoEM map of EV71 mature viron in complex with the Fab fragment of antibody D5 | | Descriptor: | Capsid protein VP1, Heavy chain of Fab fragment variable region of antibody D5, Light chain of Fab fragment variable region of antibody D5 | | Authors: | Fan, C, Ye, X.H, Ku, Z.Q, Zuo, T, Kong, L.L, Zhang, C, Shi, J.P, Liu, Q.W, Chen, T, Zhang, Y.Y, Jiang, W, Zhang, L.Q, Huang, Z, Cong, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural Basis for Recognition of Human Enterovirus 71 by a Bivalent Broadly Neutralizing Monoclonal Antibody
Plos Pathog., 12, 2016
|
|
3ZWN
 
 | | Crystal structure of Aplysia cyclase complexed with substrate NGD and product cGDPR | | Descriptor: | 3-(AMINOCARBONYL)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYD ROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2- YL]PYRIDINIUM, ADP-RIBOSYL CYCLASE, CYCLIC GUANOSINE DIPHOSPHATE-RIBOSE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
6EBA
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Unoccupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-08-06 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.812 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
6E8J
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Occupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
7QFT
 
 | | Crystal structure of KLK6 in complex with compound 16a | | Descriptor: | KLK6 Activity-Based Probe (Ahx-DPhe-Cha-Dht-Arg-DPP), Kallikrein-6 | | Authors: | Jagtap, P.K.A, Zhang, L, De Vita, E, Tate, E.W, Hennig, J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A KLK6 Activity-Based Probe Reveals a Role for KLK6 Activity in Pancreatic Cancer Cell Invasion.
J.Am.Chem.Soc., 144, 2022
|
|
6E9C
 
 | | Selenomethionine Derivative Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
6M54
 
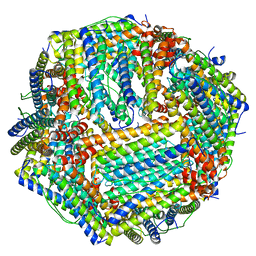 | | Human apo ferritin frozen on TEM grid with Amorphous nickel titanium alloy supporting film | | Descriptor: | FE (II) ION, Ferritin heavy chain | | Authors: | Huang, X, Zhang, L, Wen, Z, Chen, H, Li, S, Ji, G, Yin, C, Sun, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation.
Prog.Biophys.Mol.Biol., 156, 2020
|
|
4WNA
 
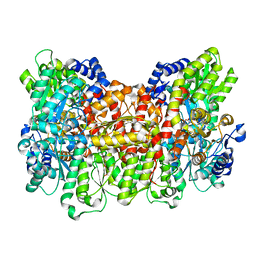 | | Structure of the Nitrogenase MoFe Protein from Azotobacter vinelandii Pressurized with Xenon | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Hoy, J.A, Zhang, L, Einsle, O, Rees, D.C. | | Deposit date: | 2014-10-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.
Biochemistry, 54, 2015
|
|
2RJG
 
 | | Crystal structure of biosynthetic alaine racemase from Escherichia coli | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
2RJH
 
 | | Crystal structure of biosynthetic alaine racemase in D-cycloserine-bound form from Escherichia coli | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
