7W59
 
 | | The cryo-EM structure of human pre-C*-I complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
4GJT
 
 | | complex structure of nectin-4 bound to MV-H | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein, Poliovirus receptor-related protein 4 | | Authors: | Zhang, X, Lu, G, Qi, J, Li, Y, He, Y, Xu, X, Shi, J, Zhang, C, Yan, J, Gao, G.F. | | Deposit date: | 2012-08-10 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4
Nat.Struct.Mol.Biol., 20, 2013
|
|
8X2I
 
 | |
8X2H
 
 | |
5HWL
 
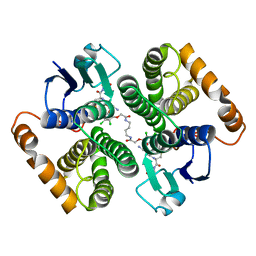 | | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 2, N,N'-(butane-1,4-diyl)bis{2-[2,3-dichloro-4-(2-methylidenebutanoyl)phenoxy]acetamide} | | Authors: | Zhang, X, Wei, J, Wu, S, Zhang, H.P, Luo, M, Yang, X.L, Liao, F, Wang, D.Q. | | Deposit date: | 2016-01-29 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form
To Be Published
|
|
7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | Descriptor: | D-Cysteine desulfhydrase | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | Descriptor: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
1ML9
 
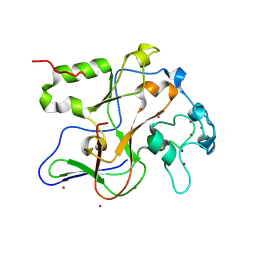 | | Structure of the Neurospora SET domain protein DIM-5, a histone lysine methyltransferase | | Descriptor: | Histone H3 methyltransferase DIM-5, UNKNOWN, ZINC ION | | Authors: | Zhang, X, Tamaru, H, Khan, S.I, Horton, J.R, Keefe, L.J, Selker, E.U, Cheng, X. | | Deposit date: | 2002-08-30 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Neurospora SET domain protein DIM-5,
a histone H3 lysine methyltransferase
Cell(Cambridge,Mass.), 111, 2002
|
|
2RF9
 
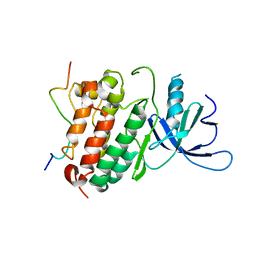 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2RFE
 
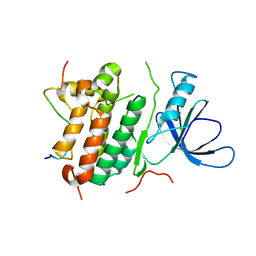 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2GS6
 
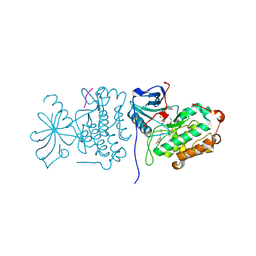 | | Crystal Structure of the active EGFR kinase domain in complex with an ATP analog-peptide conjugate | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, Peptide, ... | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Allosteric Mechanism for Activation of the Kinase Domain of Epidermal Growth Factor Receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
4GJE
 
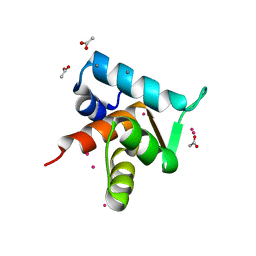 | |
4GJF
 
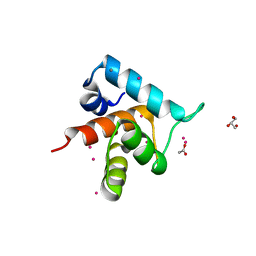 | |
4GJG
 
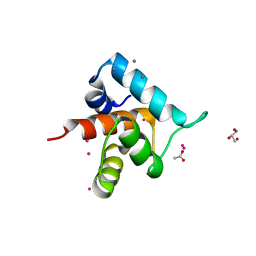 | |
8XJM
 
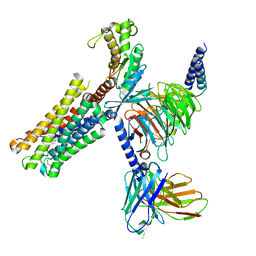 | | Latanoprost acid bound Prostaglandin F2-alpha receptor-Gq Protein Complex | | Descriptor: | Antibody fragment scFv16, Engineered miniGq, Fusion tag,Prostaglandin F2-alpha receptor,LgBiT, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
1F3L
 
 | |
1J04
 
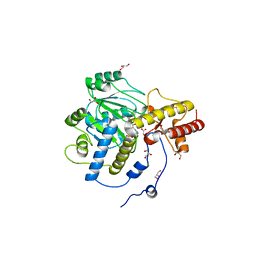 | | Structural mechanism of enzyme mistargeting in hereditary kidney stone disease in vitro | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, alanine--glyoxylate aminotransferase | | Authors: | Zhang, X, Djordjevic, S, Bartlam, M, Ye, S, Rao, Z, Danpure, C.J. | | Deposit date: | 2002-10-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural implications of a G170R mutation of alanine:glyoxylate aminotransferase that is associated with peroxisome-to-mitochondrion mistargeting.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1KJS
 
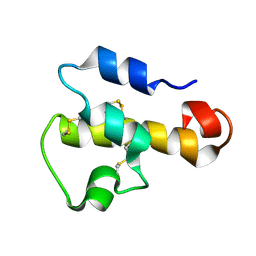 | | NMR SOLUTION STRUCTURE OF C5A AT PH 5.2, 303K, 20 STRUCTURES | | Descriptor: | C5A | | Authors: | Zhang, X, Boyar, W, Toth, M, Wennogle, L, Gonnella, N.C. | | Deposit date: | 1997-01-09 | | Release date: | 1997-05-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural definition of the C5a C terminus by two-dimensional nuclear magnetic resonance spectroscopy.
Proteins, 28, 1997
|
|
1NPU
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 | | Descriptor: | Programmed cell death protein 1 | | Authors: | Zhang, X, Schwartz, J.-C.D, Guo, X, Cao, E, Chen, L, Zhang, Z.-Y, Nathenson, S.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-20 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the costimulatory receptor programmed death-1.
Immunity, 20, 2004
|
|
1H0C
 
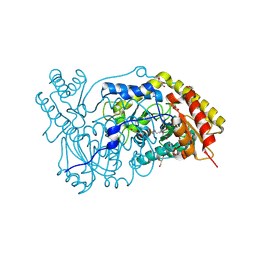 | | The crystal structure of human alanine:glyoxylate aminotransferase | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Zhang, X, Danpure, C.J, Roe, S.M, Pearl, L.H. | | Deposit date: | 2002-06-17 | | Release date: | 2003-06-12 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alanine:Glyoxylate Aminotransferase and the Relationship between Genotype and Enzymatic Phenotype in Primary Hyperoxaluria Type 1.
J.Mol.Biol., 331, 2003
|
|
1OR8
 
 | | Structure of the Predominant protein arginine methyltransferase PRMT1 | | Descriptor: | GLYCEROL, Protein arginine N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhang, X, Cheng, X. | | Deposit date: | 2003-03-12 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 and Analysis of Its Binding to Substrate Peptides
Structure, 11, 2003
|
|
1ORH
 
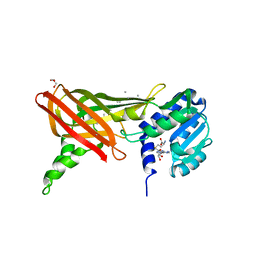 | | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 | | Descriptor: | GLYCEROL, Protein arginine N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhang, X, Cheng, X. | | Deposit date: | 2003-03-13 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 and Analysis of Its Binding to Substrate Peptides
Structure, 11, 2003
|
|
1PEG
 
 | | Structural basis for the product specificity of histone lysine methyltransferases | | Descriptor: | Histone H3, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Zhang, X, Yang, Z, Khan, S.I, Horton, J.R, Tamaru, H, Selker, E.U, Cheng, X. | | Deposit date: | 2003-05-21 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for the product specificity of histone lysine methyltransferases
Mol.Cell, 12, 2003
|
|
1NCN
 
 | |
1ORI
 
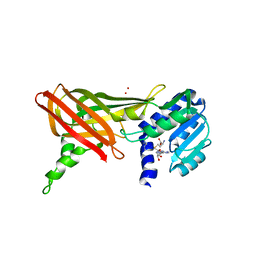 | | Structure of the predominant protein arginine methyltransferase PRMT1 | | Descriptor: | Protein arginine N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN LIGAND | | Authors: | Zhang, X, Cheng, X. | | Deposit date: | 2003-03-13 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 and Analysis of its Binding to Substrate Peptides
Structure, 11, 2003
|
|
