3J4F
 
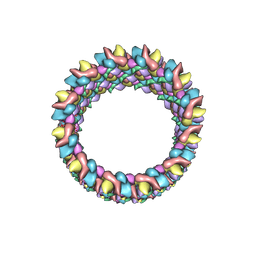 | | Structure of HIV-1 capsid protein by cryo-EM | | 分子名称: | capsid protein | | 著者 | Zhao, G, Perilla, J.R, Meng, X, Schulten, K, Zhang, P. | | 登録日 | 2013-07-11 | | 公開日 | 2013-07-24 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (8.6 Å) | | 主引用文献 | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
8FAC
 
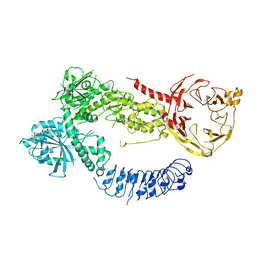 | |
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | 分子名称: | capsid protein | | 著者 | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | 登録日 | 2013-02-23 | | 公開日 | 2013-05-29 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (8.6 Å) | | 主引用文献 | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
6LZ3
 
 | |
6LZ7
 
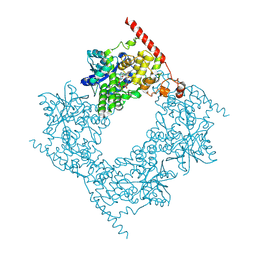 | |
2M6B
 
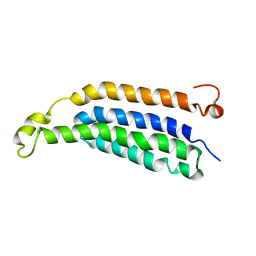 | | Structure of full-length transmembrane domains of human glycine receptor alpha1 monomer subunit | | 分子名称: | Full-Length Transmembrane Domains of Human Glycine Receptor alpha1 Subunit | | 著者 | Mowrey, D, Cui, T, Jia, Y, Ma, D, Makhov, A.M, Zhang, P, Tang, P, Xu, Y. | | 登録日 | 2013-03-28 | | 公開日 | 2013-09-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Open-Channel Structures of the Human Glycine Receptor alpha 1 Full-Length Transmembrane Domain.
Structure, 21, 2013
|
|
5E8L
 
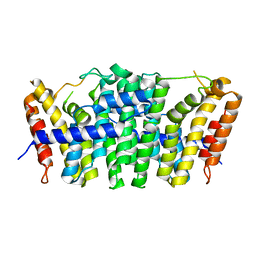 | | Crystal structure of geranylgeranyl pyrophosphate synthase 11 from Arabidopsis thaliana | | 分子名称: | Heterodimeric geranylgeranyl pyrophosphate synthase large subunit 1, chloroplastic | | 著者 | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | 登録日 | 2015-10-14 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.807 Å) | | 主引用文献 | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
5E8H
 
 | | Crystal structure of geranylfarnesyl pyrophosphate synthases 2 from Arabidopsis thaliana | | 分子名称: | Geranylgeranyl pyrophosphate synthase 3, chloroplastic | | 著者 | Wang, C, Chen, Q, Wang, G, Zhang, P. | | 登録日 | 2015-10-14 | | 公開日 | 2015-11-11 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
5E8K
 
 | | Crystal structure of polyprenyl pyrophosphate synthase 2 from Arabidopsis thaliana | | 分子名称: | Geranylgeranyl pyrophosphate synthase 10, mitochondrial | | 著者 | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | 登録日 | 2015-10-14 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.028 Å) | | 主引用文献 | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
6KZ8
 
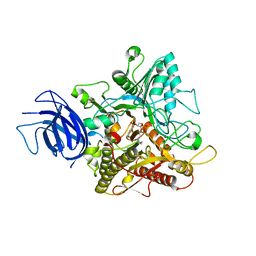 | | Crystal structure of plant Phospholipase D alpha complex with phosphatidic acid | | 分子名称: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, CALCIUM ION, Phospholipase D alpha 1 | | 著者 | Li, J.X, Yu, F, Zhang, P. | | 登録日 | 2019-09-23 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.291 Å) | | 主引用文献 | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
6KZ9
 
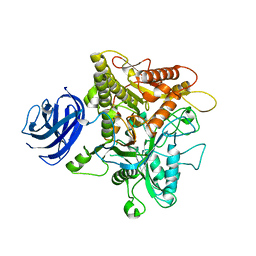 | |
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | 分子名称: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | 著者 | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | 登録日 | 2015-10-07 | | 公開日 | 2016-03-16 | | 最終更新日 | 2017-08-23 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
7VM8
 
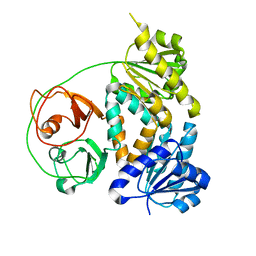 | |
7LKI
 
 | |
3IWP
 
 | |
1XRF
 
 | | The Crystal Structure of a Novel, Latent Dihydroorotase from Aquifex aeolicus at 1.7 A resolution | | 分子名称: | Dihydroorotase, SULFATE ION, ZINC ION | | 著者 | Martin, P.D, Purcarea, C, Zhang, P, Vaishnav, A, Sadecki, S, Guy-Evans, H.I, Evans, D.R, Edwards, B.F. | | 登録日 | 2004-10-14 | | 公開日 | 2005-07-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | The crystal structure of a novel, latent dihydroorotase from Aquifex aeolicus at 1.7A resolution
J.Mol.Biol., 348, 2005
|
|
3J3Y
 
 | |
6SKN
 
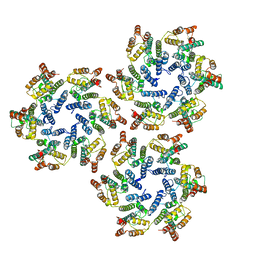 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | 分子名称: | Gag protein | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2019-08-16 | | 公開日 | 2020-08-26 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4OV4
 
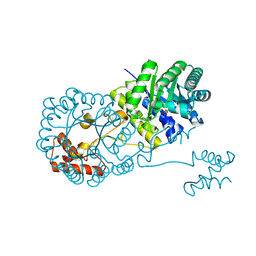 | | Isopropylmalate synthase binding with ketoisovalerate | | 分子名称: | 2-isopropylmalate synthase, 3-METHYL-2-OXOBUTANOIC ACID, ZINC ION | | 著者 | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | 登録日 | 2014-02-20 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
5XSJ
 
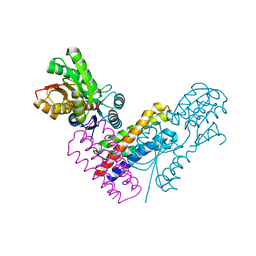 | | XylFII-LytSN complex | | 分子名称: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS, ... | | 著者 | Li, J.X, Wang, C.Y, Zhang, P. | | 登録日 | 2017-06-14 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XSD
 
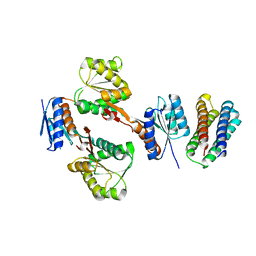 | | XylFII-LytSN complex mutant - D103A | | 分子名称: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS | | 著者 | Li, J.X, Wang, C.Y, Zhang, P. | | 登録日 | 2017-06-13 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6SKK
 
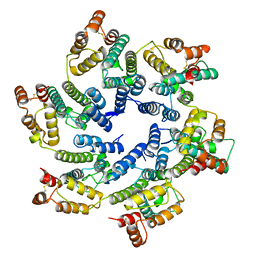 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | 分子名称: | capsid protein | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2019-08-15 | | 公開日 | 2020-08-26 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4O1I
 
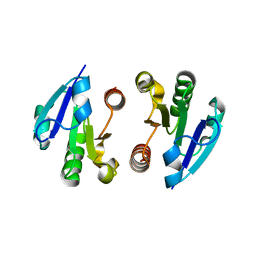 | | Crystal Structure of the regulatory domain of MtbGlnR | | 分子名称: | Transcriptional regulatory protein | | 著者 | Lin, W, Wang, C, Zhang, P. | | 登録日 | 2013-12-16 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
4O1H
 
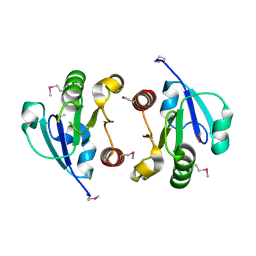 | | Crystal Structure of the regulatory domain of AmeGlnR | | 分子名称: | Transcription regulator GlnR | | 著者 | Lin, W, Wang, C, Zhang, P. | | 登録日 | 2013-12-16 | | 公開日 | 2014-04-23 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
4OV9
 
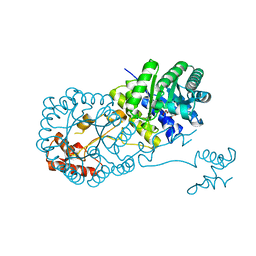 | | Structure of isopropylmalate synthase binding with alpha-isopropylmalate | | 分子名称: | (2S)-2-hydroxy-2-(propan-2-yl)butanedioic acid, ZINC ION, isopropylmalate synthase | | 著者 | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | 登録日 | 2014-02-20 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
