7X8C
 
 | | Crystal structure of a KTSC family protein from Euryarchaeon Methanolobus vulcani | | Descriptor: | KTSC domain-containing protein, SODIUM ION | | Authors: | Zhang, Z.F, Zhu, K.L, Chen, Y.Y, Cao, P, Gong, Y. | | Deposit date: | 2022-03-12 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Biochemical and structural characterization of a KTSC family single-stranded DNA-binding protein from Euryarchaea.
Int.J.Biol.Macromol., 216, 2022
|
|
3OVG
 
 | | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound | | Descriptor: | PHOSPHATE ION, ZINC ION, amidohydrolase | | Authors: | Zhang, Z, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound
To be Published
|
|
3MSR
 
 | |
7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
5W3X
 
 | | Crystal structure of PopP2 in complex with IP6, AcCoA and the WRKY domain of RRS1-R . | | Descriptor: | ACETYL COENZYME *A, Disease resistance protein RRS1, GLYCEROL, ... | | Authors: | Zhang, Z.M, Gao, L, Song, J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of host substrate acetylation by a YopJ family effector.
Nat Plants, 3, 2017
|
|
6LR7
 
 | |
3U5T
 
 | | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase from Sinorhizobium meliloti | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase from Sinorhizobium meliloti
To be Published
|
|
8HGW
 
 | | Crystal structure of MehpH in complex with MBP | | Descriptor: | 1-BUTANOL, Monoalkyl phthalate hydrolase, PHTHALIC ACID | | Authors: | Zhang, Z.M, Wang, Y.J, Chen, Y.B. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.80001163 Å) | | Cite: | Molecular insights into the catalytic mechanism of plasticizer degradation by a monoalkyl phthalate hydrolase.
Commun Chem, 6, 2023
|
|
8HGV
 
 | |
3MOW
 
 | | Crystal structure of SHP2 in complex with a tautomycetin analog TTN D-1 | | Descriptor: | (2Z)-2-[(1R)-3-{[(1R,2S,3R,6S,7S,10S,12S,15E,17E)-18-carboxy-16-ethyl-3,7-dihydroxy-1,2,6,10,12-pentamethyl-5-oxooctade ca-15,17-dien-1-yl]oxy}-1-hydroxy-3-oxopropyl]-3-methylbut-2-enedioic acid, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.-Y, Liu, S, Yu, Z, Yu, X. | | Deposit date: | 2010-04-23 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SHP2 in complex with a tautomycetin analog TTN D-1
To be Published
|
|
6LVX
 
 | | Crystal structure of TLR7/Cpd-1 (SM-374527) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-butoxy-9-(phenylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
8Q7N
 
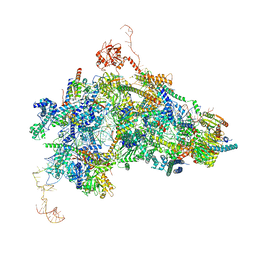 | | cryo-EM structure of the human spliceosomal B complex protomer (tri-snRNP core region) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, MINX pre-mRNA, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2023-08-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM analyses of dimerized spliceosomes provide new insights into the functions of B complex proteins.
Embo J., 43, 2024
|
|
7VJL
 
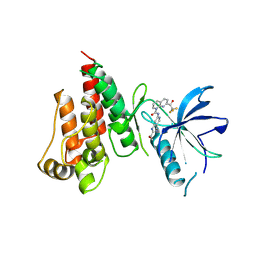 | | The crystal structure of FGFR4 kinase domain in complex with N-(5-cyano-4-((2-methoxyethyl)amino)pyridin-2-yl)-7-(2,2,2-trifluoroacetyl)-3,4-dihydro-1,8-naphthyridine-1(2H)-carboxamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[5-(aminomethyl)-4-(2-methoxyethylamino)pyridin-2-yl]-7-[2,2,2-tris(fluoranyl)ethanoyl]-3,4-dihydro-2H-1,8-naphthyridine-1-carboxamide | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.900173 Å) | | Cite: | Characterization of an aromatic trifluoromethyl ketone as a new warhead for covalently reversible kinase inhibitor design.
Bioorg.Med.Chem., 50, 2021
|
|
6JIO
 
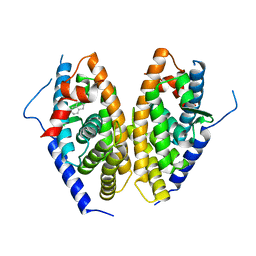 | | Human LXR-beta in complex with a ligand | | Descriptor: | Oxysterols receptor LXR-beta, tert-butyl 7-amino-3,4-dihydroisoquinoline-2(1H)-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-02-22 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identify liver X receptor beta modulator building blocks by developing a fluorescence polarization-based competition assay.
Eur.J.Med.Chem., 178, 2019
|
|
6K10
 
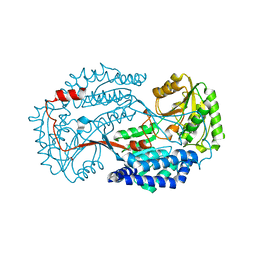 | | Non substrate bound state of Staphylococcus Aureus AldH | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase | | Authors: | Zhang, Z, Tao, X. | | Deposit date: | 2019-05-08 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78962183 Å) | | Cite: | Structural Insight into the Substrate Gating Mechanism by Staphylococcus aureus Aldehyde Dehydrogenase
CCS Chemistry, 2, 2020
|
|
7XAF
 
 | | The crystal structure of TrkA kinase in complex with 4^6,14-dimethyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-10-oxo-5-oxa-11,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclo- tetradecaphan-2-yne-45-carboxamide | | Descriptor: | 4^6,14-dimethyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-10-oxo-5-oxa-11,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclo-tetradecaphan-2-yne-45-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2022-03-17 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001182 Å) | | Cite: | Discovery of the First Highly Selective and Broadly Effective Macrocycle-Based Type II TRK Inhibitors that Overcome Clinically Acquired Resistance.
J.Med.Chem., 65, 2022
|
|
8GUP
 
 | | Crystal structure of EsaG from Staphylococcus aureus | | Descriptor: | CITRIC ACID, Type VII secretion system protein EsaG | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2022-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.298725 Å) | | Cite: | A toxin-deformation dependent inhibition mechanism in the T7SS toxin-antitoxin system of Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
8J61
 
 | | The crystal structure of TrkA kinase in complex with 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05065274 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8J63
 
 | | The crystal structure of TrkA kinase in complex with 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-11-oxo-5-oxa-10,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-11-oxo-5-oxa-10,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.0005 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8YXT
 
 | | Crystal structure of PtmB | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PtmB | | Authors: | Zhang, Z.Y, Qu, X.D, Duan, B.R, Wei, G.Z. | | Deposit date: | 2024-04-02 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A nucleobase-driven P450 peroxidase system enables regio- and stereo-specific formation of C─C and C─N bonds.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5WRO
 
 | | Crystal structure of Drosophila enolase | | Descriptor: | CADMIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Zhang, Z, Shi, Z. | | Deposit date: | 2016-12-02 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Crystal structure of enolase from Drosophila melanogaster.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
8YY7
 
 | | Crystal structure of PtmB in complex with cyclo-(L-Trp-L-Trp) and Hypoxanthine | | Descriptor: | (3S,6S)-3,6-bis[(1H-indol-3-yl)methyl]piperazine-2,5-dione, HYPOXANTHINE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z.Y, Qu, X.D, Duan, B.R, Wei, G.Z. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A nucleobase-driven P450 peroxidase system enables regio- and stereo-specific formation of C─C and C─N bonds.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8YZ8
 
 | | Crystal structure of PtmB in complex with Adenine | | Descriptor: | ADENINE, PROTOPORPHYRIN IX CONTAINING FE, PtmB | | Authors: | Zhang, Z.Y, Qu, X.D, Duan, B.R, Wei, G.Z. | | Deposit date: | 2024-04-06 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | A nucleobase-driven P450 peroxidase system enables regio- and stereo-specific formation of C─C and C─N bonds.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8YYP
 
 | | Crystal structure of PtmB in complex with cyclo-(L-Trp-L-Trp) and Adenine | | Descriptor: | (3S,6S)-3,6-bis[(1H-indol-3-yl)methyl]piperazine-2,5-dione, ADENINE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z.Y, Qu, X.D, Duan, B.R, Wei, G.Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A nucleobase-driven P450 peroxidase system enables regio- and stereo-specific formation of C─C and C─N bonds.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8YZA
 
 | | Crystal structure of PtmB in complex with cyclo-(L-Trp-L-Trp) and Guanine | | Descriptor: | (3S,6S)-3,6-bis[(1H-indol-3-yl)methyl]piperazine-2,5-dione, GUANINE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z.Y, Qu, X.D, Duan, B.R, Wei, G.Z. | | Deposit date: | 2024-04-06 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A nucleobase-driven P450 peroxidase system enables regio- and stereo-specific formation of C─C and C─N bonds.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
