1BVM
 
 | | SOLUTION NMR STRUCTURE OF BOVINE PANCREATIC PHOSPHOLIPASE A2, 20 STRUCTURES | | Descriptor: | PROTEIN (PHOSPHOLIPASE A2) | | Authors: | Yuan, C.-H, Byeon, I.-J.L, Li, Y, Tsai, M.-D. | | Deposit date: | 1998-09-14 | | Release date: | 1999-09-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of phospholipase A2 from functional perspective. 1. Functionally relevant solution structure and roles of the hydrogen-bonding network.
Biochemistry, 38, 1999
|
|
1TR4
 
 | | Solution structure of human oncogenic protein gankyrin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10 | | Authors: | Yuan, C, Li, J, Mahajan, A, Poi, M.J, Byeon, I.J, Tsai, M.D. | | Deposit date: | 2004-06-19 | | Release date: | 2004-11-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human oncogenic protein gankyrin containing seven ankyrin repeats and analysis of its structure--function relationship.
Biochemistry, 43, 2004
|
|
1K3N
 
 | | NMR Structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T155) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1K3Q
 
 | | NMR structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T192) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1J4Q
 
 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T192) PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1J4P
 
 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T155) PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1D9S
 
 | | TUMOR SUPPRESSOR P15(INK4B) STRUCTURE BY COMPARATIVE MODELING AND NMR DATA | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR B | | Authors: | Yuan, C, Ji, L, Selby, T.L, Byeon, I.J.L, Tsai, M.D. | | Deposit date: | 1999-10-29 | | Release date: | 2000-07-28 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: comparisons of conformational properties between p16(INK4A) and p18(INK4C).
J.Mol.Biol., 294, 1999
|
|
5EW6
 
 | | Structure of ligand binding region of uPARAP at pH 7.4 without calcium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type mannose receptor 2, ... | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding
Biochem.J., 473, 2016
|
|
1K3J
 
 | | Refined NMR Structure of the FHA1 Domain of Yeast Rad53 | | Descriptor: | Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1G3G
 
 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF YEAST RAD53 | | Descriptor: | PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Liao, H, Su, M, Yongkiettrakul, S, Byeon, I.-J.L, Tsai, M.-D. | | Deposit date: | 2000-10-24 | | Release date: | 2001-01-10 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structure of the FHA1 domain of yeast Rad53 and identification of binding sites for both FHA1 and its target protein Rad9
J.Mol.Biol., 304, 2000
|
|
1J4O
 
 | | REFINED NMR STRUCTURE OF THE FHA1 DOMAIN OF YEAST RAD53 | | Descriptor: | PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
3P8F
 
 | |
3P8G
 
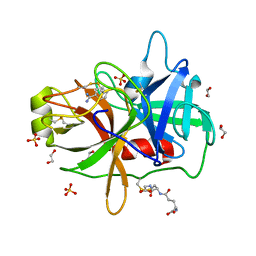 | | Crystal Structure of MT-SP1 in complex with benzamidine | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, GLUTATHIONE, ... | | Authors: | Yuan, C, Huang, M, Chen, L. | | Deposit date: | 2010-10-13 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of catalytic domain of Matriptase in complex with Sunflower trypsin inhibitor-1.
Bmc Struct.Biol., 11, 2011
|
|
6ITE
 
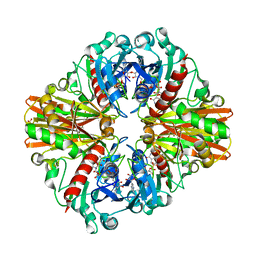 | | Crystal structure of group A Streptococcal surface dehydrogenase (SDH) | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Yuan, C, Li, R, Huang, M.D. | | Deposit date: | 2018-11-21 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structural determination of group A Streptococcal surface dehydrogenase and characterization of its interaction with urokinase-type plasminogen activator receptor.
Biochem.Biophys.Res.Commun., 510, 2019
|
|
5E4K
 
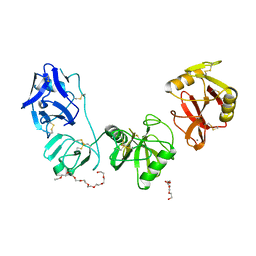 | | Structure of ligand binding region of uPARAP at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, C-type mannose receptor 2, ... | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2015-10-06 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding
Biochem.J., 473, 2016
|
|
5E4L
 
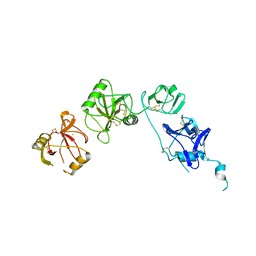 | | Structure of ligand binding region of uPARAP at pH 5.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type mannose receptor 2, CALCIUM ION | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2015-10-06 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of uPARAP, a member of mannose receptor family
to be published
|
|
2JQL
 
 | | NMR structure of the yeast Dun1 FHA domain in complex with a doubly phosphorylated (pT) peptide derived from Rad53 SCD1 | | Descriptor: | DNA damage response protein kinase DUN1, Serine/threonine-protein kinase RAD53 | | Authors: | Yuan, C, Lee, H, Chang, C, Heierhorst, J, Tsai, M. | | Deposit date: | 2007-06-02 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Diphosphothreonine-specific interaction between an SQ/TQ cluster and an FHA domain in the Rad53-Dun1 kinase cascade.
Mol.Cell, 30, 2008
|
|
2JQJ
 
 | | NMR structure of yeast Dun1 FHA domain | | Descriptor: | DNA damage response protein kinase DUN1 | | Authors: | Yuan, C, Lee, H, Chang, C, Heierhorst, J, Tsai, M. | | Deposit date: | 2007-06-02 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Diphosphothreonine-specific interaction between an SQ/TQ cluster and an FHA domain in the Rad53-Dun1 kinase cascade.
Mol.Cell, 30, 2008
|
|
2JQI
 
 | |
7V63
 
 | | Structure of dimeric uPAR at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2021-08-19 | | Release date: | 2021-12-22 | | Last modified: | 2022-04-06 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Crystal structure and cellular functions of uPAR dimer
Nat Commun, 13, 2022
|
|
4K23
 
 | | Structure of anti-uPAR Fab ATN-658 | | Descriptor: | anti-uPAR antibody, heavy chain, light chain | | Authors: | Yuan, C, Huang, M, Chen, L. | | Deposit date: | 2013-04-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a New Epitope in uPAR as a Target for the Cancer Therapeutic Monoclonal Antibody ATN-658, a Structural Homolog of the uPAR Binding Integrin CD11b ( alpha M)
Plos One, 9, 2014
|
|
1A7A
 
 | | STRUCTURE OF HUMAN PLACENTAL S-ADENOSYLHOMOCYSTEINE HYDROLASE: DETERMINATION OF A 30 SELENIUM ATOM SUBSTRUCTURE FROM DATA AT A SINGLE WAVELENGTH | | Descriptor: | (1'R,2'S)-9-(2-HYDROXY-3'-KETO-CYCLOPENTEN-1-YL)ADENINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYLHOMOCYSTEINE HYDROLASE | | Authors: | Turner, M.A, Yuan, C.-S, Borchardt, R.T, Hershfield, M.S, Smith, G.D, Howell, P.L. | | Deposit date: | 1998-03-10 | | Release date: | 1999-04-20 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength.
Nat.Struct.Biol., 5, 1998
|
|
4XSK
 
 | | Structure of PAItrap, an uPA mutant | | Descriptor: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PAItrap, an uPA mutant
To Be Published
|
|
1LXC
 
 | | Crystal Structure of E. Coli Enoyl Reductase-NAD+ with a Bound Acrylamide Inhibitor | | Descriptor: | 3-(6-AMINOPYRIDIN-3-YL)-N-METHYL-N-[(1-METHYL-1H-INDOL-2-YL)METHYL]ACRYLAMIDE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miller, W.H, Seefeld, M.A, Newlander, K.A, Uzinskas, I.N, Burgess, W.J, Heerding, D.A, Yuan, C.C.K, Head, M.S, Payne, D.J, Rittenhouse, S.F, Moore, T.D, Pearson, S.C, Dewolf, V, Berry, W.E, Keller, P.M, Polizzi, B.J, Qiu, X, Janson, C.A, Huffman, W.F. | | Deposit date: | 2002-06-05 | | Release date: | 2002-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of aminopyridine-based inhibitors of bacterial enoyl-ACP reductase (FabI).
J.Med.Chem., 45, 2002
|
|
2FA9
 
 | | The crystal structure of Sar1[H79G]-GDP provides insight into the coat-controlled GTP hydrolysis in the disassembly of COP II | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rao, Y, Huang, M, Yuan, C, Bian, C, Hou, X. | | Deposit date: | 2005-12-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Sar1[H79G]-GDP Which Provides Insight into the Coat-controlled GTP Hydrolysis in the Disassembly of COP II
Chin.J.Struct.Chem., 25, 2006
|
|
