2KPU
 
 | | NMR Structure of YbbR family protein Dhaf_0833 (residues 32-118) from Desulfitobacterium hafniense DCB-2: Northeast Structural Genomics Consortium target DhR29B | | Descriptor: | YbbR family protein | | Authors: | Cort, J.R, Ramelot, T.A, Yang, Y, Belote, R.L, Ciccosanti, C, Haleema, J, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structures of domains I and IV from YbbR are representative of a widely distributed protein family.
Protein Sci., 20, 2011
|
|
7QUR
 
 | | SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry | | Descriptor: | 2,3,5-tris(iodanyl)benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7QUS
 
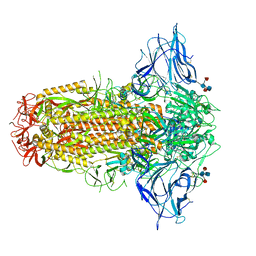 | | SARS-CoV-2 Spike, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-08 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
3TCL
 
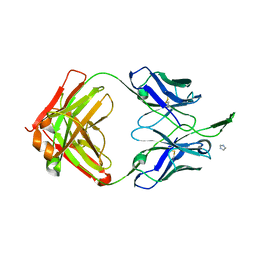 | | Crystal Structure of HIV-1 Neutralizing Antibody CH04 | | Descriptor: | CH04 Heavy Chain Fab, CH04 Light Chain Fab, IMIDAZOLE | | Authors: | Louder, R.K, McLellan, J.S, Pancera, M, Yang, Y, Zhang, B, Bonsignori, M, Kwong, P.D. | | Deposit date: | 2011-08-09 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
2EEM
 
 | |
1G47
 
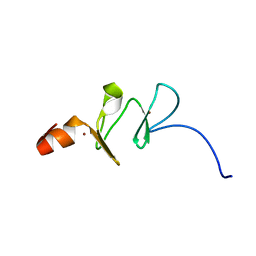 | | 1ST LIM DOMAIN OF PINCH PROTEIN | | Descriptor: | PINCH PROTEIN, ZINC ION | | Authors: | Velyvis, A, Yang, Y, Wu, C, Qin, J. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the focal adhesion adaptor PINCH LIM1 domain and characterization of its interaction with the integrin-linked kinase ankyrin repeat domain.
J.Biol.Chem., 276, 2001
|
|
1D5Q
 
 | | SOLUTION STRUCTURE OF A MINI-PROTEIN REPRODUCING THE CORE OF THE CD4 SURFACE INTERACTING WITH THE HIV-1 ENVELOPE GLYCOPROTEIN | | Descriptor: | CHIMERIC MINI-PROTEIN | | Authors: | Vita, C, Drakopoulou, E, Vizzanova, J, Rochette, S, Martin, L, Menez, A, Roumestand, C, Yang, Y.S, Ylisastigui, L, Benjouad, A, Gluckman, J.C. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6X4X
 
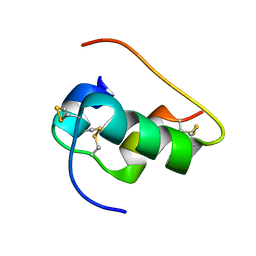 | | B24Y DKP insulin | | Descriptor: | Insulin, Insulin chain A | | Authors: | Weiss, M.A, Yang, Y. | | Deposit date: | 2020-05-24 | | Release date: | 2020-08-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Evolution of insulin at the edge of foldability and its medical implications.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XQB
 
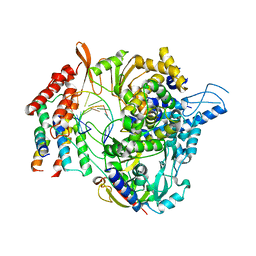 | | SARS-CoV-2 RdRp/RNA complex | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-07-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of SARS-CoV-2 RdRp/RNA complex at 3.4 Angstroms resolution
To Be Published
|
|
6XV2
 
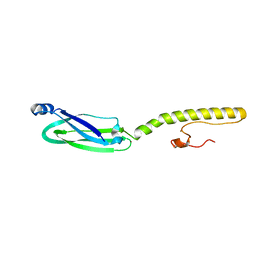 | | Full structure of RYMV P1 protein, derived from crystallographic and NMR data. | | Descriptor: | ZINC ION, p1 | | Authors: | Poignavent, V, Hoh, F, Vignols, F, Demene, H, Yang, Y, Gillet, F.X. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Flexible and Original Architecture of Two Unrelated Zinc Fingers Underlies the Role of the Multitask P1 in RYMV Spread.
J.Mol.Biol., 434, 2022
|
|
1MQ8
 
 | | Crystal structure of alphaL I domain in complex with ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-L, ... | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MQA
 
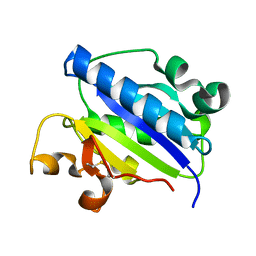 | | Crystal structure of high affinity alphaL I domain in the absence of ligand or metal | | Descriptor: | Integrin alpha-L | | Authors: | Shimaoka, T, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, Zhang, R, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | Descriptor: | Integrin alpha-L, MANGANESE (II) ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
6VI8
 
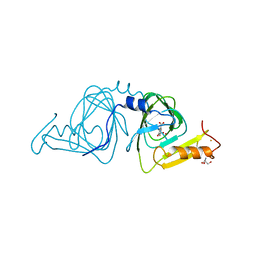 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a superoxo bound structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VIA
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a seven-membered lactone bound structure | | Descriptor: | (2R,3E)-2-hydroxy-3-imino-2,3-dihydrooxepine-4-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1NFG
 
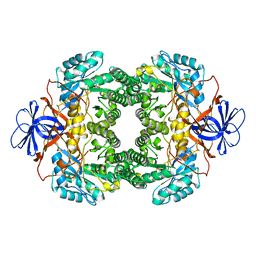 | | Structure of D-hydantoinase | | Descriptor: | D-hydantoinase, ZINC ION | | Authors: | Xu, Z, Yang, Y, Jiang, W, Arnold, E, Ding, J. | | Deposit date: | 2002-12-14 | | Release date: | 2003-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of D-Hydantoinase from Burkholderia pickettii at a Resolution of 2.7 Angstroms: Insights into the Molecular Basis of Enzyme Thermostability.
J.Bacteriol., 185, 2003
|
|
5DAY
 
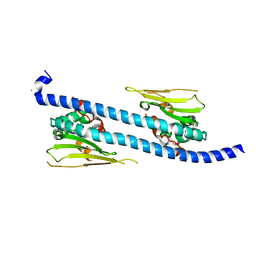 | | The structure of NAP1-Related Protein(NRP1) in Arabidopsis | | Descriptor: | CALCIUM ION, NAP1-related protein 1 | | Authors: | Zhu, Y, Rong, L, Yang, Y, Zhang, C, Feng, H.Y, Zheng, L.N, Shen, W.H, Ma, J.B, Dong, A.W. | | Deposit date: | 2015-08-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | The structure of NAP1-Related Protein(NRP1) in Arabidopsis
To Be Published
|
|
5E1J
 
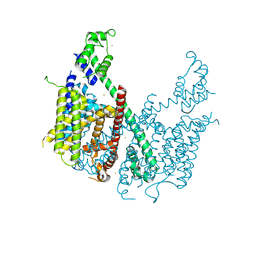 | | Structure of voltage-gated two-pore channel TPC1 from Arabidopsis thaliana | | Descriptor: | BARIUM ION, CALCIUM ION, Two pore calcium channel protein 1 | | Authors: | Guo, J, Zeng, W, Chen, Q, Lee, C, Chen, L, Yang, Y, Jiang, Y. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.308 Å) | | Cite: | Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana.
Nature, 531, 2016
|
|
5LQ6
 
 | | Salmonella effector SpvD - R161 variant | | Descriptor: | SODIUM ION, Virulence protein vsdE | | Authors: | Zhang, Y, Grabe, G.J, Rolhion, N, Yang, Y, Holden, D.W, Hare, S.A. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The Salmonella Effector SpvD Is a Cysteine Hydrolase with a Serovar-specific Polymorphism Influencing Catalytic Activity, Suppression of Immune Responses, and Bacterial Virulence.
J. Biol. Chem., 291, 2016
|
|
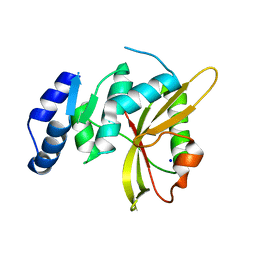
5JZV
 
 | | The structure of D77G hCINAP-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase isoenzyme 6 | | Authors: | Liu, Y, Yang, Z, Yang, Y, Cai, X, Zheng, X. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The ATPase hCINAP regulates 18S rRNA processing and is essential for embryogenesis and tumour growth.
Nat Commun, 7, 2016
|
|
3NAF
 
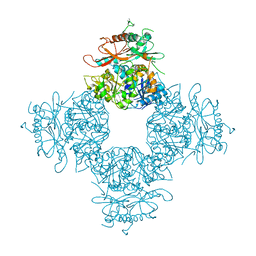 | | Structure of the Intracellular Gating Ring from the Human High-conductance Ca2+ gated K+ Channel (BK Channel) | | Descriptor: | Calcium-activated potassium channel subunit alpha-1,Calcium-activated potassium channel subunit alpha-1,Calcium-activated potassium channel subunit alpha-1 | | Authors: | Wu, Y, Yang, Y, Ye, S, Jiang, Y. | | Deposit date: | 2010-06-01 | | Release date: | 2010-06-30 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the gating ring from the human large-conductance Ca(2+)-gated K(+) channel.
Nature, 466, 2010
|
|
2JWE
 
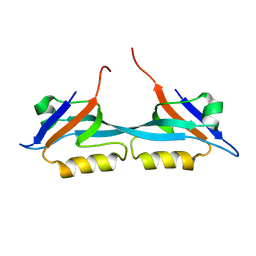 | | Solution structure of the second PDZ domain from human zonula occludens-1: A dimeric form with 3D domain swapping | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Ji, P, Wu, J.W, Zhang, J.H, Yang, Y.S, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2007-10-10 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second PDZ domain of Zonula Occludens 1
Proteins, 79, 2011
|
|
3OWT
 
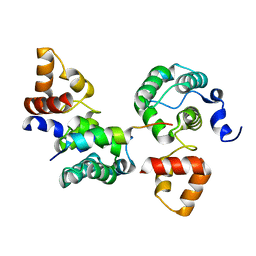 | | Crystal structure of S. cerevisiae RAP1-Sir3 complex | | Descriptor: | DNA-binding protein RAP1, Regulatory protein SIR3 | | Authors: | Chen, Y, Yang, Y, Lei, M. | | Deposit date: | 2010-09-20 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3RSN
 
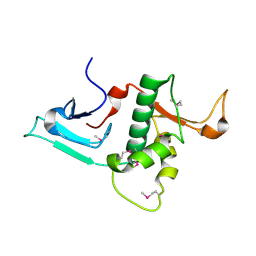 | | Crystal Structure of the N-terminal region of Human Ash2L | | Descriptor: | Set1/Ash2 histone methyltransferase complex subunit ASH2, ZINC ION | | Authors: | Chen, Y, Wan, B, Wang, K.C, Cao, F, Yang, Y, Protacio, A, Dou, Y, Chang, H.Y, Lei, M. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding.
Embo Rep., 12, 2011
|
|
5EZK
 
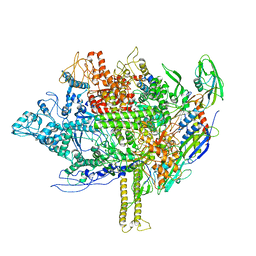 | | RNA polymerase model placed by Molecular replacement into X-ray diffraction map of DNA-bound RNA Polymerase-Sigma 54 holoenzyme complex. | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darbari, V.C, Yang, Y, Lu, D, Zhang, N, Glyde, R, Wang, Y, Murakami, K.S, Buck, M, Zhang, X. | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.5 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase- Sigma 54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
