7XE0
 
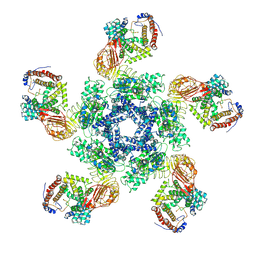 | | Cryo-EM structure of plant NLR Sr35 resistosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AvrSr35, Sr35 | | Authors: | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
6JKY
 
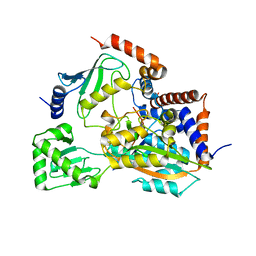 | |
6K1T
 
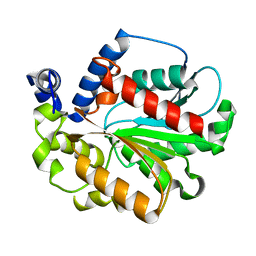 | |
6K4R
 
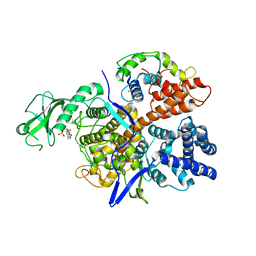 | |
6K11
 
 | |
6K4K
 
 | |
6K4L
 
 | |
6KUE
 
 | |
4DKS
 
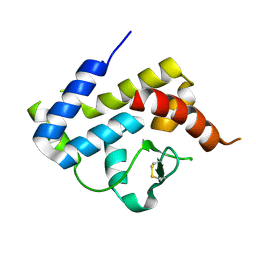 | | A spindle-shaped virus protein (chymotrypsin treated) | | Descriptor: | Probable integrase | | Authors: | Ouyang, S, Liang, W, Huang, L, Liu, Z.-J. | | Deposit date: | 2012-02-04 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of the C-terminal catalytic domain of SSV1 integrase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6KWY
 
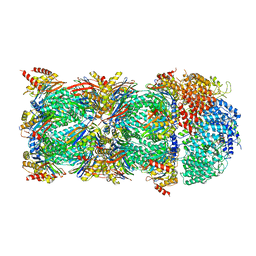 | | human PA200-20S complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Proteasome activator complex subunit 4, Proteasome subunit alpha type-1, ... | | Authors: | Ouyang, S, Hongxin, G. | | Deposit date: | 2019-09-09 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Plos Biol., 18, 2020
|
|
5LSC
 
 | | The structure of the metallo-beta-lactamase VIM-2 in complex with a triazolylthioacetamide inhibitor | | Descriptor: | 2-[5-[2-(1,3-benzothiazol-2-ylamino)-2-oxidanylidene-ethyl]sulfanyl-4~{H}-1,2,4-triazol-3-yl]benzoic acid, CHLORIDE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Christopeit, T, Yang, K.-W, Yang, S.-K, Leiros, H.-K.S. | | Deposit date: | 2016-08-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | The structure of the metallo-beta-lactamase VIM-2 in complex with a triazolylthioacetamide inhibitor.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
7MYN
 
 | | Cryo-EM Structure of p110alpha in complex with p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1QNA
 
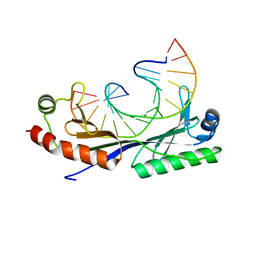 | | Crystal structure of the T(-30) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*AP*AP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN7
 
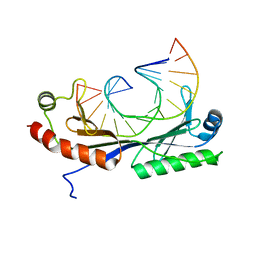 | | Crystal structure of the T(-27) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*TP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*AP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN3
 
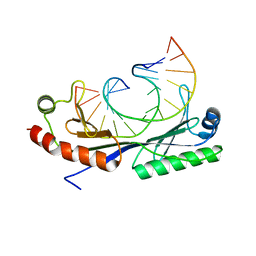 | | Crystal structure of the C(-25) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*CP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*GP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN5
 
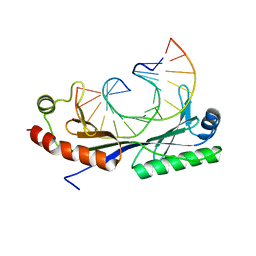 | | Crystal structure of the G(-26) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*GP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*CP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
4AEF
 
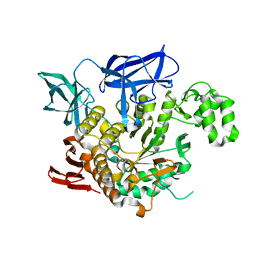 | | THE CRYSTAL STRUCTURE OF THERMOSTABLE AMYLASE FROM THE PYROCOCCUS | | Descriptor: | NEOPULLULANASE (ALPHA-AMYLASE II) | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Yang, S.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2012-01-10 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A Novel Domain Arrangement in a Monomeric Cyclodextrin-Hydrolyzing Enzyme from the Hyperthermophile Pyrococcus Furiosus.
Biochim.Biophys.Acta, 1834, 2013
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XMK
 
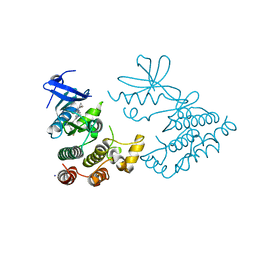 | | Crystal structure of human RIPK1 kinase domain in complex with compound SKLB923 | | Descriptor: | 5-[2-(cyclopropylcarbonylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-(3-fluorophenyl)ethyl]-1-methyl-indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Zhang, L, Wang, Y, Li, Y, Yang, S. | | Deposit date: | 2022-04-26 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.376 Å) | | Cite: | From Hit to Lead: Structure-Based Optimization of Novel Selective Inhibitors of Receptor-Interacting Protein Kinase 1 (RIPK1) for the Treatment of Inflammatory Diseases.
J.Med.Chem., 67, 2024
|
|
5WPR
 
 | | Crystal structure HpiC1 in C2 space group | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WPP
 
 | | Crystal structure HpiC1 W73M/K132M | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
