8F28
 
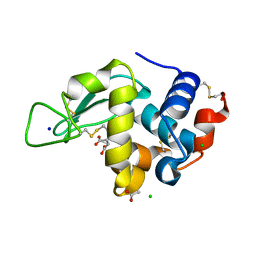 | | Lysozyme Structures from Single-Entity Crystallization Method NanoAC | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Yang, R, Sankaran, B, Ogbonna, E, Wang, G. | | Deposit date: | 2022-11-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Single-Entity Method for Actively Controlled Nucleation and High-Quality Protein Crystal Synthesis.
Anal.Chem., 95, 2023
|
|
5H0I
 
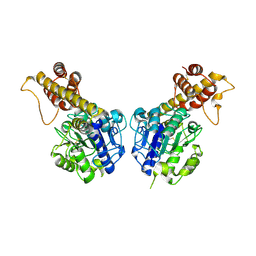 | |
9CAQ
 
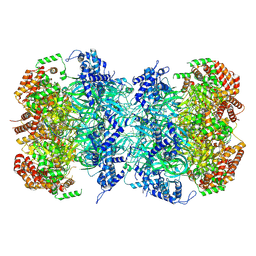 | | Cryo-EM structure of a human MCM2-7 double hexamer formed from independently loaded MCM2-7 single hexamers | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (44-MER), ... | | Authors: | Yang, R, Hunker, O, Bleichert, F. | | Deposit date: | 2024-06-17 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Multiple mechanisms for licensing human replication origins.
Nature, 636, 2024
|
|
8G5V
 
 | | Empty capsid of Hepatitis B virus | | Descriptor: | Core protein Cp183 | | Authors: | Yang, R, Cingolani, G. | | Deposit date: | 2023-02-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for nuclear import of hepatitis B virus (HBV) nucleocapsid core.
Sci Adv, 10, 2024
|
|
8G6V
 
 | |
8G8Y
 
 | |
8W0I
 
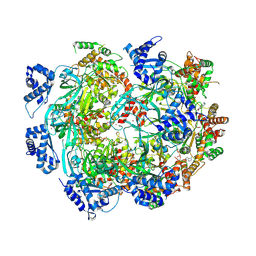 | | Cryo-EM structure of the human MCM2-7 heterohexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA replication licensing factor MCM2, ... | | Authors: | Yang, R, Hunker, O, Bleichert, F. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Multiple mechanisms for licensing human replication origins.
Nature, 636, 2024
|
|
8W0G
 
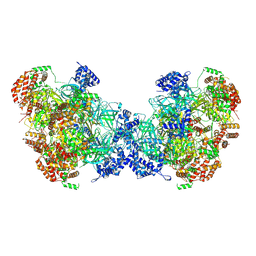 | | Cryo-EM structure of a human MCM2-7 dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA replication licensing factor MCM2, ... | | Authors: | Yang, R, Hunker, O, Bleichert, F. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Multiple mechanisms for licensing human replication origins.
Nature, 636, 2024
|
|
7UMI
 
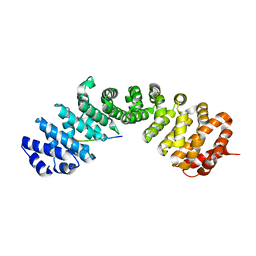 | | Importin a1 bound to Cp183-CTD | | Descriptor: | HBV-NLS, Importin subunit alpha-1 | | Authors: | Yang, R, Cingolani, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for nuclear import of hepatitis B virus (HBV) nucleocapsid core.
Sci Adv, 10, 2024
|
|
7UCJ
 
 | | Mammalian 80S translation initiation complex with mRNA and Harringtonine | | Descriptor: | 18S rRNA, 28s rRNA, 40S ribosomal protein S10, ... | | Authors: | Yang, R, Arango, D, Sturgill, D, Oberdoerffer, S. | | Deposit date: | 2022-03-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine.
Mol.Cell, 82, 2022
|
|
7UCK
 
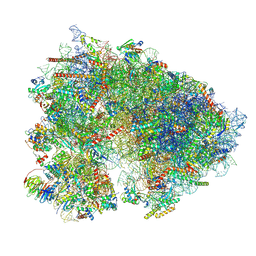 | | 80S translation initiation complex with ac4c(-1) mRNA and Harringtonine | | Descriptor: | 18S rRNA, 28s rRNA, 40S ribosomal protein S10, ... | | Authors: | Yang, R, Arango, D, Sturgill, D, Oberdoerffer, S. | | Deposit date: | 2022-03-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine.
Mol.Cell, 82, 2022
|
|
8AU1
 
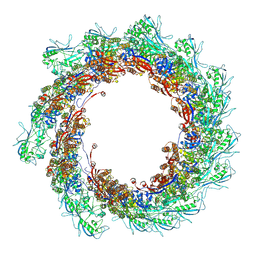 | | Jumbo Phage phi-kp24 tail outer sheath | | Descriptor: | Putative tail sheath protein | | Authors: | Ouyang, R, Briegel, A. | | Deposit date: | 2022-08-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
8BFK
 
 | | Jumbo Phage phi-kp24 tail inner tube | | Descriptor: | Putative virion structural protein | | Authors: | Ouyang, R, Briegel, A. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
8BFL
 
 | | Jumbo Phage phi-kp24 empty capsid hexamers | | Descriptor: | Major head protein | | Authors: | Ouyang, R. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
8BFP
 
 | | Jumbo Phage phi-kp24 empty capsid pentamer hexamers | | Descriptor: | Major head protein | | Authors: | Ouyang, R. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
2OC8
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH503034 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C virus, ZINC ION, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R.S, Arasappan, A, Bennett, F, Bogen, S.L, Chen, K, Jao, E, Liu, Y.T, Lovey, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
9HFS
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): hexamer with dCTP bound in the active site | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Activity and structure of human (d)CTP deaminase CDADC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9HFQ
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): trimer without a ligand | | Descriptor: | Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Activity and structure of human (d)CTP deaminase CDADC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9HFT
 
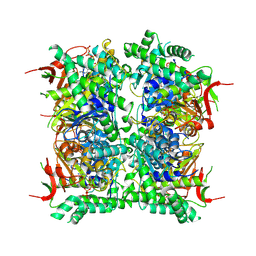 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): hexamer with 5mdCTP bound in the active site | | Descriptor: | Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION, [[(2~{R},3~{S},5~{R})-5-(4-azanyl-5-methyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activity and structure of human (d)CTP deaminase CDADC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9HFR
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): hexamer without a ligand | | Descriptor: | Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Activity and structure of human (d)CTP deaminase CDADC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8Y13
 
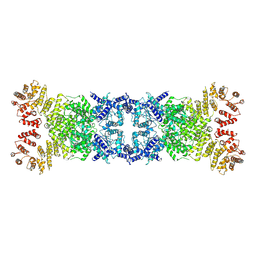 | | Cryo-EM structure of anti-phage defense associated DSR2 tetramer (H171A) | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Li, F.X, Shi, Z.B, Wang, R.W, Xu, Q, Yang, R, Wu, Z.X. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
6BZ9
 
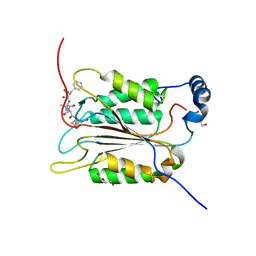 | | Crystal structure of human caspase-1 in complex with Ac-FLTD-CMK | | Descriptor: | Ac-FLTD-CMK, Caspase-1, DI(HYDROXYETHYL)ETHER | | Authors: | Xiao, T.S, Yang, J, Liu, Z, Wang, C, Yang, R. | | Deposit date: | 2017-12-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1MWN
 
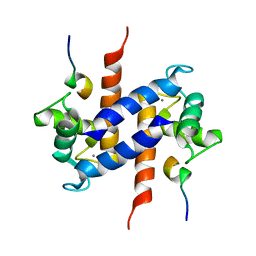 | | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12 | | Descriptor: | CALCIUM ION, F-actin capping protein alpha-1 subunit, S-100 protein, ... | | Authors: | Inman, K.G, Yang, R, Rustandi, R.R, Miller, K.E, Baldisseri, D.M, Weber, D.J. | | Deposit date: | 2002-09-30 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12
J.Mol.Biol., 324, 2002
|
|
7SP4
 
 | | In situ cryo-EM structure of bacteriophage Sf6 gp3:gp7:gp5 complex in conformation 2 at 3.71A resolution | | Descriptor: | Gene 3 protein, Gene 5 protein, Gene 7 protein | | Authors: | Li, F, Cingolani, G, Hou, C, Yang, R. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | High-resolution cryo-EM structure of the Shigella virus Sf6 genome delivery tail machine.
Sci Adv, 8, 2022
|
|
7SPU
 
 | | In situ cryo-EM structure of bacteriophage Sf6 gp3:gp7:gp5 complex in conformation 1 at 3.73A resolution | | Descriptor: | Gene 3 protein, Gene 5 protein, Gene 7 protein | | Authors: | Li, F, Cingolani, G, Hou, C, Yang, R. | | Deposit date: | 2021-11-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | High-resolution cryo-EM structure of the Shigella virus Sf6 genome delivery tail machine.
Sci Adv, 8, 2022
|
|
