2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC7
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH571696 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, TERT-BUTYL {(1S)-2-[(1R,2S,5R)-2-({[(1S)-3-AMINO-1-(CYCLOBUTYLMETHYL)-2,3-DIOXOPROPYL]AMINO}CARBONYL)-7,7-DIMETHYL-6-OXA-3-AZABICYCLO[3.2.0]HEPT-3-YL]-1-CYCLOHEXYL-2-OXOETHYL}CARBAMATE, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
1MWN
 
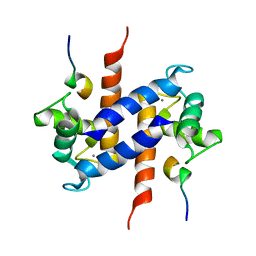 | | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12 | | Descriptor: | CALCIUM ION, F-actin capping protein alpha-1 subunit, S-100 protein, ... | | Authors: | Inman, K.G, Yang, R, Rustandi, R.R, Miller, K.E, Baldisseri, D.M, Weber, D.J. | | Deposit date: | 2002-09-30 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12
J.Mol.Biol., 324, 2002
|
|
2OC8
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH503034 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C virus, ZINC ION, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R.S, Arasappan, A, Bennett, F, Bogen, S.L, Chen, K, Jao, E, Liu, Y.T, Lovey, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
8WLO
 
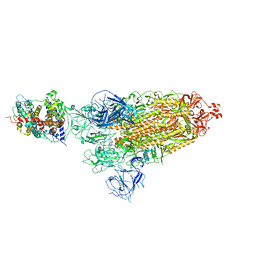 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein in complex with hippopotamus ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Yang, R.R, Li, S.H. | | Deposit date: | 2023-09-30 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Molecular basis of hippopotamus ACE2 binding to SARS-CoV-2.
J.Virol., 98, 2024
|
|
7SP4
 
 | | In situ cryo-EM structure of bacteriophage Sf6 gp3:gp7:gp5 complex in conformation 2 at 3.71A resolution | | Descriptor: | Gene 3 protein, Gene 5 protein, Gene 7 protein | | Authors: | Li, F, Cingolani, G, Hou, C, Yang, R. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | High-resolution cryo-EM structure of the Shigella virus Sf6 genome delivery tail machine.
Sci Adv, 8, 2022
|
|
7SPU
 
 | | In situ cryo-EM structure of bacteriophage Sf6 gp3:gp7:gp5 complex in conformation 1 at 3.73A resolution | | Descriptor: | Gene 3 protein, Gene 5 protein, Gene 7 protein | | Authors: | Li, F, Cingolani, G, Hou, C, Yang, R. | | Deposit date: | 2021-11-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | High-resolution cryo-EM structure of the Shigella virus Sf6 genome delivery tail machine.
Sci Adv, 8, 2022
|
|
7SZ6
 
 | | Kinetically trapped Pseudomonas-phage PaP3 portal protein - delta barrel mutant class-3 | | Descriptor: | Portal protein | | Authors: | Hou, C.-F.D, Swanson, N.A, Li, F, Yang, R, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2021-11-25 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.24 Å) | | Cite: | Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3.
J.Mol.Biol., 434, 2022
|
|
7SZ4
 
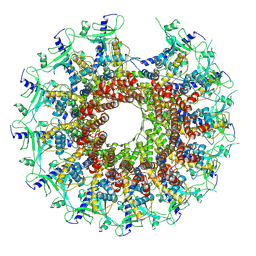 | | Kinetically trapped Pseudomonas-phage PaP3 portal protein - delta barrel mutant class-2 | | Descriptor: | Portal protein | | Authors: | Hou, C.-F.D, Swanson, N.A, Li, F, Yang, R, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2021-11-25 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3.
J.Mol.Biol., 434, 2022
|
|
7SYA
 
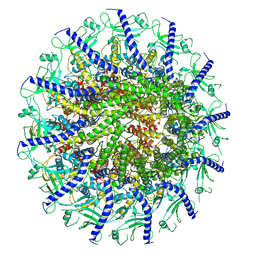 | | Kinetically trapped Pseudomonas-phage PaP3 portal protein - Full Length | | Descriptor: | Portal protein | | Authors: | Hou, C.F.D, Swanson, N.A, Li, F, Yang, R, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3.
J.Mol.Biol., 434, 2022
|
|
7SXK
 
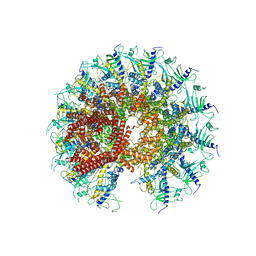 | | Kinetically trapped Pseudomonas-phage PaP3 portal protein - Full Length | | Descriptor: | Portal protein | | Authors: | Hou, C.F.D, Swanson, N.A, Li, F, Yang, R, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2021-11-23 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3.
J.Mol.Biol., 434, 2022
|
|
8WLR
 
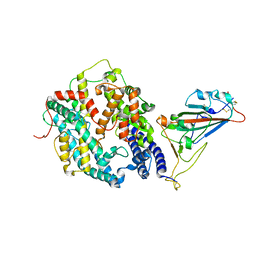 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein receptor-binding domain in complex with hippopotamus ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | Authors: | Han, P, Yang, R.R, Li, S.H. | | Deposit date: | 2023-09-30 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Molecular basis of hippopotamus ACE2 binding to SARS-CoV-2.
J.Virol., 98, 2024
|
|
5ZHB
 
 | |
8W0E
 
 | |
8W0F
 
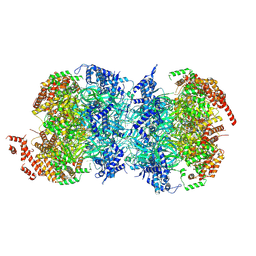 | |
5ZIG
 
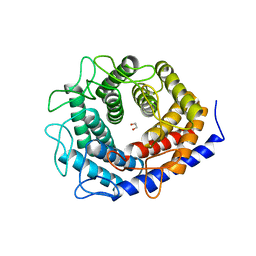 | |
7TJF
 
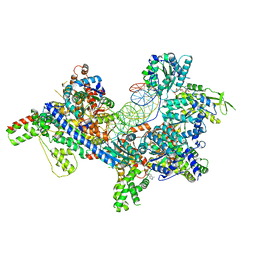 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA, 84 bp ARS1, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJI
 
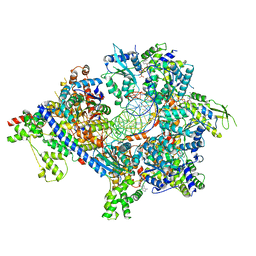 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 2) with flexible Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJH
 
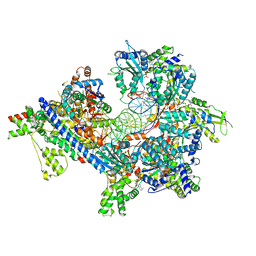 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with flexible Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJJ
 
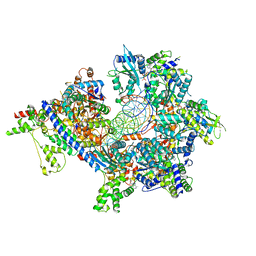 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with docked Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJK
 
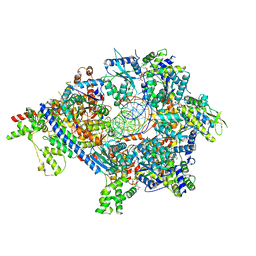 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 2) with docked Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
6BZ9
 
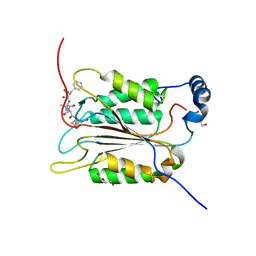 | | Crystal structure of human caspase-1 in complex with Ac-FLTD-CMK | | Descriptor: | Ac-FLTD-CMK, Caspase-1, DI(HYDROXYETHYL)ETHER | | Authors: | Xiao, T.S, Yang, J, Liu, Z, Wang, C, Yang, R. | | Deposit date: | 2017-12-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
