7YGW
 
 | | Crystal structure of the Zn2+-bound EFhd1/Swiprosin-2 | | Descriptor: | EF-hand domain-containing protein D1, GLYCEROL, ZINC ION | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
7YGY
 
 | | Crystal structure of the Zn2+-bound EFhd2/Swiprosin-1 | | Descriptor: | EF-hand domain-containing protein D2, ZINC ION | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
1GZN
 
 | | Structure of PKB kinase domain | | Descriptor: | RAC-BETA SERINE/THREONINE PROTEIN KINASE | | Authors: | Barford, D, Yang, J, Hemmings, B.A. | | Deposit date: | 2002-05-24 | | Release date: | 2003-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism for the Regulation of Protein Kinase B/Akt by Hydrophobic Motif Phosphorylation
Mol.Cell, 9, 2002
|
|
1GZO
 
 | |
1GZK
 
 | |
2APV
 
 | | Crystal Structure of the G17E/A52V/S54N/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ1
 
 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
4UI9
 
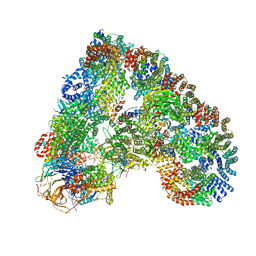 | | Atomic structure of the human Anaphase-Promoting Complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic Structure of the Apc and its Mechanism of Protein Ubiquitination
Nature, 522, 2015
|
|
2APF
 
 | | Crystal Structure of the A52V/S54N/K66E variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APT
 
 | | Crystal Structure of the G17E/S54N/K66E/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APW
 
 | | Crystal Structure of the G17E/A52V/S54N/K66E/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ3
 
 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 | | Descriptor: | Enterotoxin type C-3, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APB
 
 | | Crystal Structure of the S54N variant of murine T cell receptor Vbeta 8.2 domain | | Descriptor: | MALONIC ACID, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APX
 
 | | Crystal Structure of the G17E/A52V/S54N/K66E/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ2
 
 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, SODIUM ION, SULFATE ION, ... | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
8SSU
 
 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 19mer DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (19-MER) Strand I, DNA (19-MER) Strand II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SST
 
 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) K365T Mutant Complexed with 23mer | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSS
 
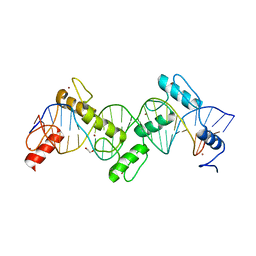 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) Complexed with 23mer | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSQ
 
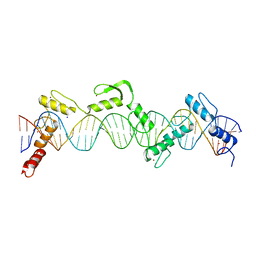 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-4 | | Descriptor: | DNA (35-MER) Strand 2, DNA (35-MER) Strand I, SODIUM ION, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSR
 
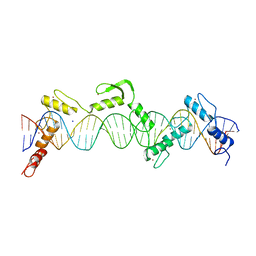 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-20 | | Descriptor: | DNA (35-MER) Strand I, DNA (35-MER) Strand II, SODIUM ION, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
6JUI
 
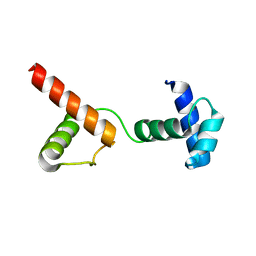 | | The atypical Myb-like protein Cdc5 contains two distinct nucleic acid-binding surfaces | | Descriptor: | Pre-mRNA-splicing factor CEF1 | | Authors: | Wang, C, Li, G, Li, M, Yang, J, Liu, J. | | Deposit date: | 2019-04-14 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Two distinct nucleic acid binding surfaces of Cdc5 regulate development.
Biochem.J., 476, 2019
|
|
7YYH
 
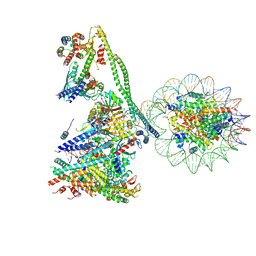 | | Structure of the human CCANdeltaT CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7YWX
 
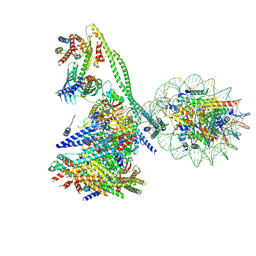 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
8A5Y
 
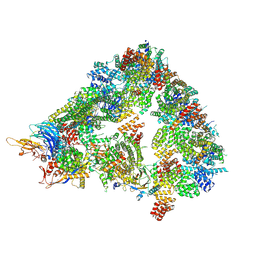 | | S. cerevisiae apo unphosphorylated APC/C. | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex and comparison to apo unphosphorylated and phosphorylated states
To Be Published
|
|
6IGB
 
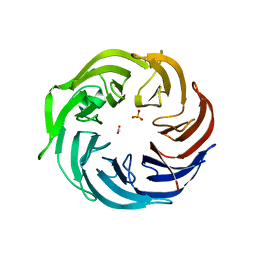 | | the structure of Pseudomonas aeruginosa Periplasmic gluconolactonase, PpgL | | Descriptor: | ACETATE ION, Periplasmic gluconolactonase, PpgL, ... | | Authors: | Song, Y.J, Shen, Y.L, Wang, K.L, Li, T, Zhu, Y.B, Li, C.C, He, L.H, Zhao, N.L, Zhao, C, Yang, J, Huang, Q, Mu, X.Y. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and Functional Insights into PpgL, a Metal-Independent beta-Propeller Gluconolactonase That Contributes toPseudomonas aeruginosaVirulence.
Infect.Immun., 87, 2019
|
|
