7CBZ
 
 | | Crystal structure of T2R-TTL-A31 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[5-[4-[2-[4-(2-cyclopropylethanoyl)piperazin-1-yl]ethoxy]phenyl]pyridin-2-yl]-N-(phenylmethyl)ethanamide, CALCIUM ION, ... | | Authors: | Yang, J.H, Yan, W. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design, Synthesis, and Bioactivity Evaluation of Dual-Target Inhibitors of Tubulin and Src Kinase Guided by Crystal Structure.
J.Med.Chem., 64, 2021
|
|
1F9S
 
 | | CRYSTAL STRUCTURE OF PLATELET FACTOR 4 MUTANT 2 | | Descriptor: | PLATELET FACTOR 4 | | Authors: | Yang, J, Doyle, M, Faulk, T, Visentin, G, Aster, R, Edwards, B. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure Comparison of Two Platelet Factor 4 Mutants with the Wild-type Reveals the Epitopes for the Heparin-induced Thrombocytopenia Antibodies
To be Published
|
|
1F9P
 
 | | CRYSTAL STRUCTURE OF CONNECTIVE TISSUE ACTIVATING PEPTIDE-III(CTAP-III) COMPLEXED WITH POLYVINYLSULFONIC ACID | | Descriptor: | CONNECTIVE TISSUE ACTIVATING PEPTIDE-III, ETHANESULFONIC ACID | | Authors: | Yang, J, Faulk, T, Aster, R, Visentin, G, Edwards, B, Castor, C. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the CXC Chemokine, Connective Tissue Activating Peptide-III, Complexed with the Heparin Analogue, Polyvinylsulfonic Acid
To be Published
|
|
1F9R
 
 | | CRYSTAL STRUCTURE OF PLATELET FACTOR 4 MUTANT 1 | | Descriptor: | PLATELET FACTOR 4 | | Authors: | Yang, J, Doyle, M, Faulk, T, Visentin, G, Aster, R, Edwards, B. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Comparison of Two Platelet Factor 4 Mutants with the Wild-type Reveals the Epitopes for the Heparin-induced Thrombocytopenia Antibodies
To be Published
|
|
1F9Q
 
 | | CRYSTAL STRUCTURE OF PLATELET FACTOR 4 | | Descriptor: | PLATELET FACTOR 4 | | Authors: | Yang, J, Doyle, M, Faulk, T, Visentin, G, Aster, R, Edwards, B. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Comparison of Two Platelet Factor 4 Mutants with the Wild-Type Reveals the Epitopes for the Heparin-Induced Thrombocytopenia Antibodies
To be Published
|
|
2RMP
 
 | | RMP-pepstatin A complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MUCOROPEPSIN, PEPSTATIN, ... | | Authors: | Yang, J, Quail, J.W. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Rhizomucor miehei aspartic proteinase complexed with the inhibitor pepstatin A at 2.7 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1UHV
 
 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | 1,5-anhydro-2-deoxy-2-fluoro-D-xylitol, Beta-xylosidase | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-11 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
2BBR
 
 | | Crystal Structure of MC159 Reveals Molecular Mechanism of DISC Assembly and vFLIP Inhibition | | Descriptor: | AZIDE ION, Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Yang, J.K, Wang, L, Zheng, L, Wan, F, Ahmed, M, Lenardo, M.J, Wu, H. | | Deposit date: | 2005-10-17 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition.
Mol.Cell, 20, 2005
|
|
2BBZ
 
 | | Crystal Structure of MC159 Reveals Molecular Mechanism of DISC Assembly and vFLIP Inhibition | | Descriptor: | Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Yang, J.K, Wang, L, Zheng, L, Wan, F, Ahmed, M, Lenardo, M.J, Wu, H. | | Deposit date: | 2005-10-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition.
Mol.Cell, 20, 2005
|
|
2WMO
 
 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
1RDQ
 
 | | Hydrolysis of ATP in the crystal of Y204A mutant of cAMP-dependent protein kinase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Ten Eyck, L.F, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-05 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a cAMP-dependent Protein Kinase Mutant at 1.26A: New Insights into the Catalytic Mechanism.
J.Mol.Biol., 336, 2004
|
|
5IFG
 
 | | Crystal structure of HigA-HigB complex from E. Coli | | Descriptor: | Antitoxin HigA, mRNA interferase HigB | | Authors: | Yang, J.S, Zhou, K, Gao, z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural insight into the E. coli HigBA complex
Biochem. Biophys. Res. Commun., 478, 2016
|
|
7N1R
 
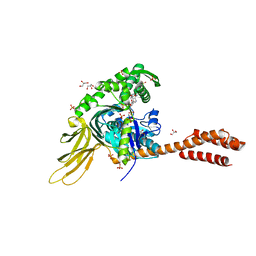 | | A novel and unique ATP hydrolysis to AMP by a human Hsp70 BiP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, Endoplasmic reticulum chaperone BiP, ... | | Authors: | Yang, J, Musayev, F, Liu, Q. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel and unique ATP hydrolysis to AMP by a human Hsp70 Binding immunoglobin protein (BiP).
Protein Sci., 31, 2022
|
|
7CUX
 
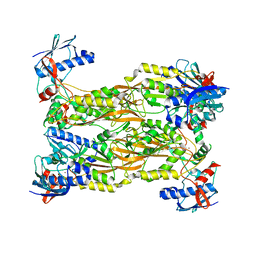 | | Crystal structure of human Schlafen 5 N'-terminal domain (SLFN5-N) involved in ssRNA cleaving and DNA binding | | Descriptor: | Schlafen family member 5, ZINC ION | | Authors: | Yang, J.Y, Luo, M, Ou, J.Y, Wang, Z.W, Gao, S. | | Deposit date: | 2020-08-25 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.29477072 Å) | | Cite: | Crystal structure of human Schlafen 5 N'-terminal domain (SLFN5-N) involved in ssRNA cleaving and DNA binding
To Be Published
|
|
1UTA
 
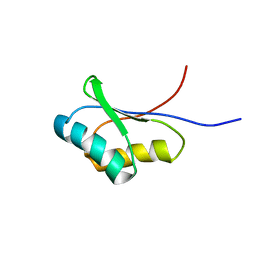 | | Solution structure of the C-terminal RNP domain from the divisome protein FtsN | | Descriptor: | CELL DIVISION PROTEIN FTSN | | Authors: | Yang, J.-C, van den Ent, F, Neuhaus, D, Brevier, J, Lowe, J. | | Deposit date: | 2003-12-04 | | Release date: | 2004-09-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Domain Architecture of the Divisome Protein Ftsn
Mol.Microbiol., 52, 2004
|
|
6PZP
 
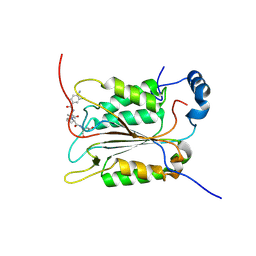 | | Crystal structure of caspase-1 in complex with VX-765 | | Descriptor: | Caspase-1, N-(4-amino-3-chlorobenzene-1-carbonyl)-3-methyl-L-valyl-N-[(2S)-1-carboxy-3-oxopropan-2-yl]-L-prolinamide | | Authors: | Yang, J, Liu, Z, Xiao, T.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of caspase-1 in complex with VX-765
To Be Published
|
|
1RE5
 
 | | Crystal structure of 3-carboxy-cis,cis-muconate lactonizing enzyme from Pseudomonas putida | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-carboxy-cis,cis-muconate cycloisomerase, CITRIC ACID | | Authors: | Yang, J, Wang, Y, Woolridge, E.M, Petsko, G.A, Kozarich, J.W, Ringe, D. | | Deposit date: | 2003-11-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of 3-Carboxy-cis,cis-muconate Lactonizing Enzyme from Pseudomonas putida, a Fumarase Class II Type Cycloisomerase: Enzyme Evolution in Parallel Pathways.
Biochemistry, 43, 2004
|
|
3QAM
 
 | | Crystal Structure of Glu208Ala mutant of catalytic subunit of cAMP-dependent protein kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | Authors: | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | Deposit date: | 2011-01-11 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
3QAL
 
 | | Crystal Structure of Arg280Ala mutant of Catalytic subunit of cAMP-dependent Protein Kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | Authors: | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | Deposit date: | 2011-01-11 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
3R4L
 
 | | Human very long half life Plasminogen Activator Inhibitor type-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasminogen activator inhibitor 1, beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, J, Zheng, H, Han, Q, Skrzypczak-Jankun, E, Jankun, J. | | Deposit date: | 2011-03-17 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Remarkable extension of PAI-1 half-life surprisingly brings no changes to its structure.
Int.J.Mol.Med., 29, 2012
|
|
3PYY
 
 | | Discovery and Characterization of a Cell-Permeable, Small-molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site | | Descriptor: | (5R)-5-[3-(4-fluorophenyl)-1-phenyl-1H-pyrazol-4-yl]imidazolidine-2,4-dione, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, GLYCEROL, ... | | Authors: | Yang, J, Campobasso, N, Biju, M.P, Fisher, K, Pan, X.Q, Cottom, J, Galbraith, S, Ho, T, Zhang, H, Hong, X, Ward, P, Hofmann, G, Siegfried, B. | | Deposit date: | 2010-12-13 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Characterization of a Cell-Permeable, Small-Molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site.
Chem.Biol., 18, 2011
|
|
2WMN
 
 | | Structure of the complex between DOCK9 and Cdc42-GDP. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
2WM9
 
 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GLYCEROL | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-06-30 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
1NFQ
 
 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Androsterone, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-15 | | Release date: | 2002-12-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFR
 
 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-16 | | Release date: | 2002-12-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
