3BTY
 
 | | Crystal structure of human ABH2 bound to dsDNA containing 1meA through cross-linking away from active site | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*DCP*DTP*DGP*DTP*DAP*DTP*(MA7)P*DAP*DCP*DTP*DGP*DCP*DG)-3'), DNA (5'-D(*DTP*DCP*DGP*DCP*DAP*DGP*DTP*DTP*DAP*DTP*DAP*DCP*DA)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
3BTZ
 
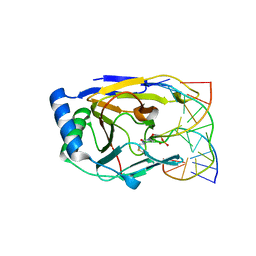 | | Crystal structure of human ABH2 cross-linked to dsDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*AP*GP*GP*TP*GP*AP*(2YR)P*AP*AP*TP*GP*CP*G)-3'), DNA (5'-D(*DTP*DCP*DGP*DCP*DAP*DTP*DTP*DAP*DTP*DCP*DAP*DCP*DC)-3') | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
3BUC
 
 | | X-ray structure of human ABH2 bound to dsDNA with Mn(II) and 2KG | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*DCP*DTP*DGP*DTP*DAP*DTP*(MA7)P*DAP*DCP*DTP*DGP*DCP*DG)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2008-01-02 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
8IT9
 
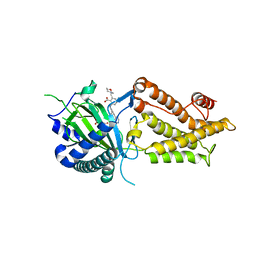 | | Co-crystal structure of FTO bound to 22 | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[(2,6-diethyl-4-pyridin-4-yl-phenyl)amino]-6-(1,4-oxazepan-4-ylmethyl)benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | Authors: | Yang, C.-G, Gan, J.H. | | Deposit date: | 2023-03-22 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Rational Design of RNA Demethylase FTO Inhibitors with Enhanced Antileukemia Drug-Like Properties.
J.Med.Chem., 66, 2023
|
|
7ML0
 
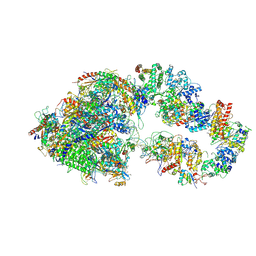 | | RNA polymerase II pre-initiation complex (PIC1) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MK9
 
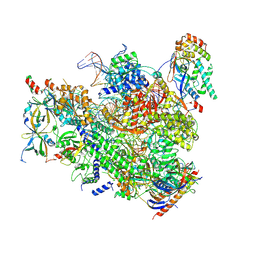 | | Complex structure of trailing EC of EC+EC (trailing EC-focused) | | Descriptor: | DNA (40-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML1
 
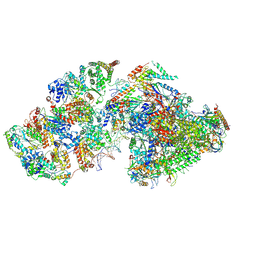 | | RNA polymerase II pre-initiation complex (PIC2) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML3
 
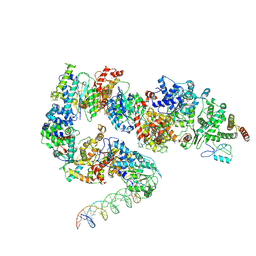 | | General transcription factor TFIIH (weak binding) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, General transcription and DNA repair factor IIH, General transcription and DNA repair factor IIH helicase subunit XPB, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML4
 
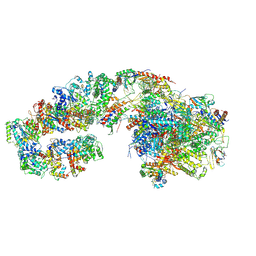 | | RNA polymerase II initially transcribing complex (ITC) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML2
 
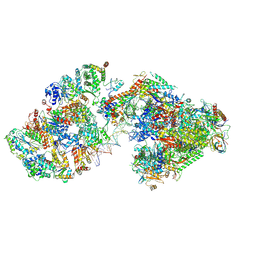 | | RNA polymerase II pre-initiation complex (PIC3) | | Descriptor: | BJ4_G0004860.mRNA.1.CDS.1, BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MKA
 
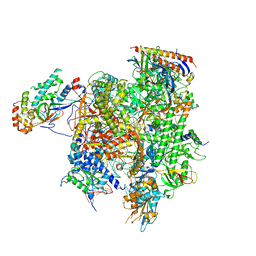 | | Structure of EC+EC (leading EC-focused) | | Descriptor: | DNA (40-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7AYE
 
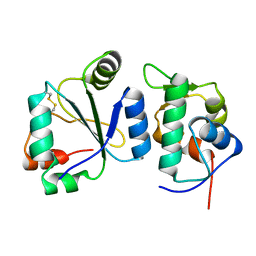 | | Crystal structure of the computationally designed chemically disruptable heterodimer LD6-MDM2 | | Descriptor: | Isoform 11 of E3 ubiquitin-protein ligase Mdm2, Thiol:disulfide interchange protein DsbD | | Authors: | Yang, C, Lau, K, Pojer, F, Correia, B.E. | | Deposit date: | 2020-11-12 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A rational blueprint for the design of chemically-controlled protein switches.
Nat Commun, 12, 2021
|
|
7MEI
 
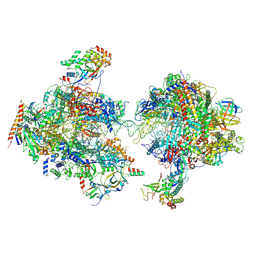 | | Composite structure of EC+EC | | Descriptor: | DNA (74-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-06 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
6YWD
 
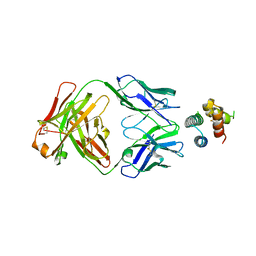 | | De novo designed protein 4H_01 in complex with Mota antibody | | Descriptor: | Antibody Mota, Heavy Chain, Light Chain, ... | | Authors: | Yang, C, Sesterhenn, F, Pojer, F, Correia, B.E. | | Deposit date: | 2020-04-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Bottom-up de novo design of functional proteins with complex structural features.
Nat.Chem.Biol., 17, 2021
|
|
6YWC
 
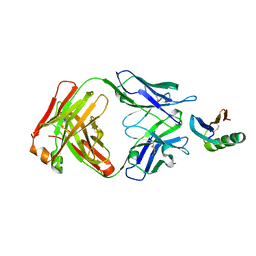 | | De novo designed protein 4E1H_95 in complex with 101F antibody | | Descriptor: | Antibody 101F, Heavy Chain, light chain, ... | | Authors: | Yang, C, Sesterhenn, F, Pojer, F, Correia, B.E. | | Deposit date: | 2020-04-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bottom-up de novo design of functional proteins with complex structural features.
Nat.Chem.Biol., 17, 2021
|
|
6IHU
 
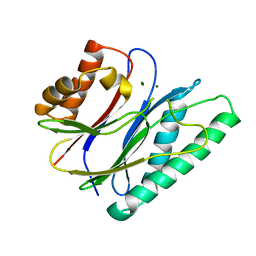 | |
1SQI
 
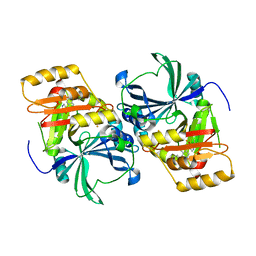 | | Structural basis for inhibitor selectivity revealed by crystal structures of plant and mammalian 4-hydroxyphenylpyruvate dioxygenases | | Descriptor: | (1-TERT-BUTYL-5-HYDROXY-1H-PYRAZOL-4-YL)[6-(METHYLSULFONYL)-4'-METHOXY-2-METHYL-1,1'-BIPHENYL-3-YL]METHANONE, 4-hydroxyphenylpyruvic acid dioxygenase, FE (III) ION | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
6IHS
 
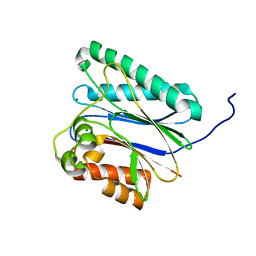 | |
6IHR
 
 | |
6IHV
 
 | |
6XXV
 
 | | Crystal Structure of a computationally designed Immunogen S2_1.2 in complex with its elicited antibody C57 | | Descriptor: | Antibody C57, Heavy Chain, Light Chain, ... | | Authors: | Yang, C, Sesterhenn, F, Correia, B.E, Pojer, F. | | Deposit date: | 2020-01-28 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.20116425 Å) | | Cite: | De novo protein design enables the precise induction of RSV-neutralizing antibodies.
Science, 368, 2020
|
|
6IHT
 
 | |
6IHL
 
 | | Crystal structure of bacterial serine phosphatase | | Descriptor: | MAGNESIUM ION, Phosphorylated protein phosphatase | | Authors: | Yang, C.-G, yang, T. | | Deposit date: | 2018-09-30 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Structural Insight into the Mechanism of Staphylococcus aureus Stp1 Phosphatase.
Acs Infect Dis., 5, 2019
|
|
6IHW
 
 | |
1SQD
 
 | | Structural basis for inhibitor selectivity revealed by crystal structures of plant and mammalian 4-hydroxyphenylpyruvate dioxygenases | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
