7VNB
 
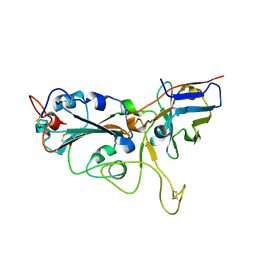 | | Crystal structure of the SARS-CoV-2 RBD in complex with a human single domain antibody n3113 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, n3113 | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
2LUH
 
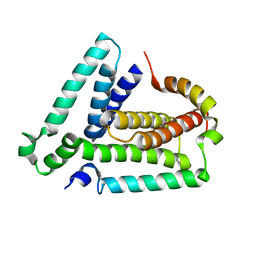 | | NMR structure of the Vta1-Vps60 complex | | Descriptor: | Vacuolar protein sorting-associated protein VTA1, Vacuolar protein-sorting-associated protein 60 | | Authors: | Yang, Z, Vild, C, Ju, J, Zhang, X, Liu, J, Shen, J, Zhao, B, Lan, W, Gong, F, Liu, M, Cao, C, Xu, Z. | | Deposit date: | 2012-06-13 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Molecular Recognition between ESCRT-III-like Protein Vps60 and AAA-ATPase Regulator Vta1 in the Multivesicular Body Pathway.
J.Biol.Chem., 287, 2012
|
|
8IMW
 
 | |
6U0N
 
 | |
7TLE
 
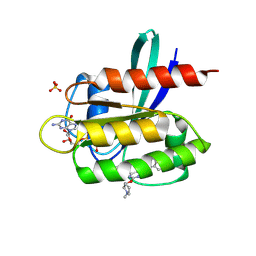 | | Crystal Structure of small molecule beta-lactone 1 covalently bound to K-Ras(G12S) | | Descriptor: | (3R,4R)-1-[7-(8-chloronaphthalen-1-yl)-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]-3-hydroxypiperidine-4-carbaldehyde, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ziyang, Z, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98716748 Å) | | Cite: | Chemical acylation of an acquired serine suppresses oncogenic signaling of K-Ras(G12S).
Nat.Chem.Biol., 18, 2022
|
|
7TLK
 
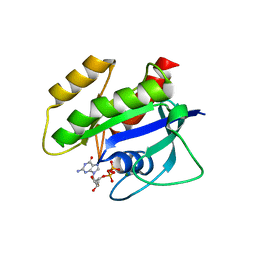 | | Crystal Structure of K-Ras(G12S) | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Ziyang, Z, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71102226 Å) | | Cite: | Chemical acylation of an acquired serine suppresses oncogenic signaling of K-Ras(G12S).
Nat.Chem.Biol., 18, 2022
|
|
7TLG
 
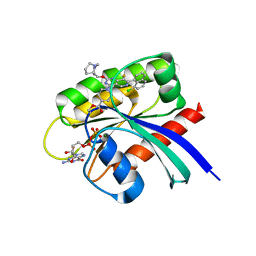 | | Crystal Structure of small molecule beta-lactone 5 covalently bound to K-Ras(G12S) | | Descriptor: | (3R,4R)-1-[7-(8-chloronaphthalen-1-yl)-8-fluoro-2-{[(4S,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-4-yl]-3-hydroxypiperidine-4-carbaldehyde, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ziyang, Z, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.80000722 Å) | | Cite: | Chemical acylation of an acquired serine suppresses oncogenic signaling of K-Ras(G12S).
Nat.Chem.Biol., 18, 2022
|
|
5HDT
 
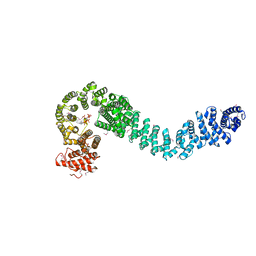 | | Human cohesin regulator Pds5B bound to a Wapl peptide | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Sister chromatid cohesion protein PDS5 homolog B, Wings apart-like protein homolog | | Authors: | Ouyang, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2016-01-05 | | Release date: | 2016-03-09 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Structure of the human cohesin regulator Pds5 in complex with Wapl motif
To Be Published
|
|
3K6J
 
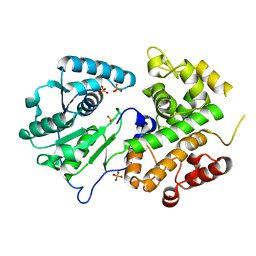 | | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans | | Descriptor: | PHOSPHATE ION, Protein F01G10.3, confirmed by transcript evidence, ... | | Authors: | Ouyang, Z, Zhang, K, Zhai, Y, Lu, J, Sun, F. | | Deposit date: | 2009-10-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans
To be Published
|
|
7CCG
 
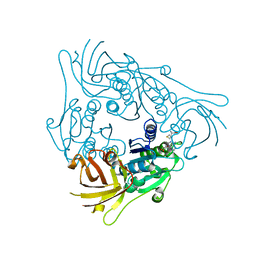 | |
9C57
 
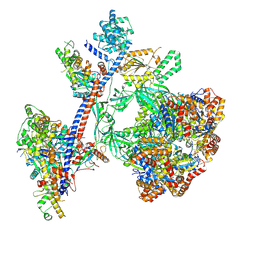 | | Reconstituted P400 Subcomplex of the human TIP60 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-05 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
9C4B
 
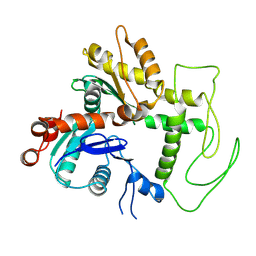 | | Second BAF53a of the human TIP60 complex | | Descriptor: | Actin-like protein 6A | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
9C47
 
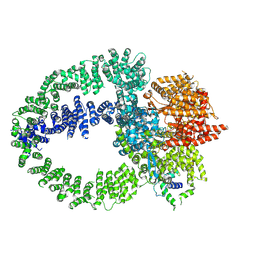 | | TRRAP module of the human TIP60 complex | | Descriptor: | E1A-binding protein p400, Transformation/transcription domain-associated protein | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
9C6N
 
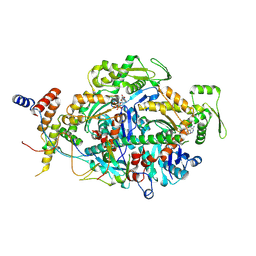 | | ARP module of the human TIP60 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-07 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
9C62
 
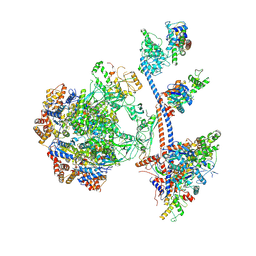 | | P400 subcomplex of the native human TIP60 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-07 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (5.28 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
4WJA
 
 | | Crystal Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
3ECN
 
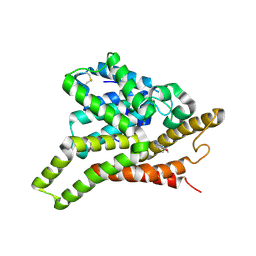 | | Crystal structure of PDE8A catalytic domain in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
3ECM
 
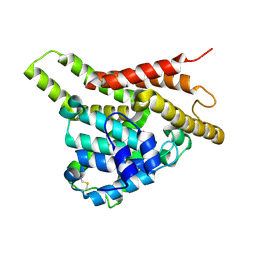 | | Crystal structure of the unliganded PDE8A catalytic domain | | Descriptor: | High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ZINC ION | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
4DNW
 
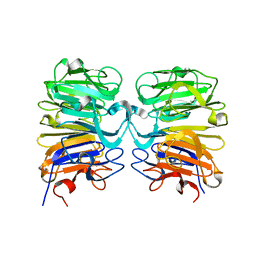 | | Crystal structure of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNV
 
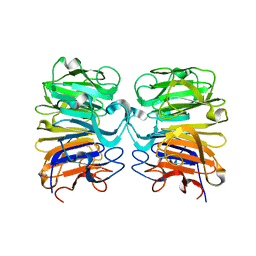 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
5JHH
 
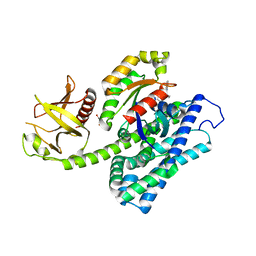 | | Crystal structure of the ternary complex between the human RhoA, its inhibitor and the DH/PH domain of human ARHGEF11 | | Descriptor: | 3-{3-[ethyl(quinolin-2-yl)amino]phenyl}propanoic acid, GLYCEROL, Rho guanine nucleotide exchange factor 11, ... | | Authors: | Lv, Z, Wang, R, Ma, L, Miao, Q, Wu, J, Yan, Z, Li, J, Miao, L, Wang, F. | | Deposit date: | 2016-04-21 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization and preliminary X-ray crystallographic analysis of a small GTPase RhoA bound with its inhibitor and PDZRhoGEF
To Be Published
|
|
9IUK
 
 | | The structure of Candida albicans Cdr1 in apo state | | Descriptor: | Pip2(20:4/18:0), Pleiotropic ABC efflux transporter of multiple drugs CDR1 | | Authors: | Peng, Y, Sun, H, Yan, Z.F. | | Deposit date: | 2024-07-22 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms.
Nat Commun, 15, 2024
|
|
9IUL
 
 | | The structure of Candida albicans Cdr1 in fluconazole-bound state | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Pip2(20:4/18:0), Pleiotropic ABC efflux transporter of multiple drugs CDR1 | | Authors: | Peng, Y, Sun, H, Yan, Z.F. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms.
Nat Commun, 15, 2024
|
|
9IUM
 
 | |
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
