6N7N
 
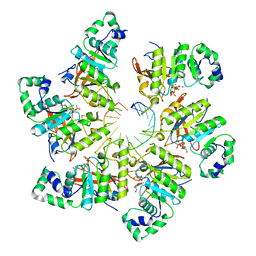 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (form I) | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA primase/helicase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | Deposit date: | 2018-11-27 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6N9W
 
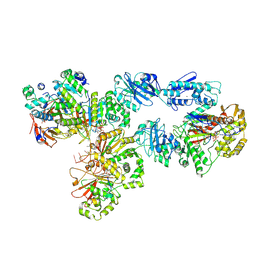 | | Structure of bacteriophage T7 lagging-strand DNA polymerase (D5A/E7A) and gp4 (helicase/primase) bound to DNA including RNA/DNA hybrid, and an incoming dTTP (LagS2) | | Descriptor: | DNA primase/helicase, DNA-directed DNA polymerase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6N7S
 
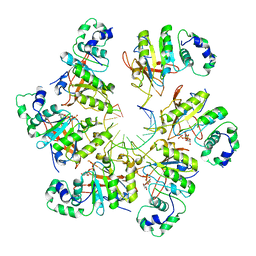 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (form II) | | Descriptor: | DNA (25-MER), DNA primase/helicase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6N7T
 
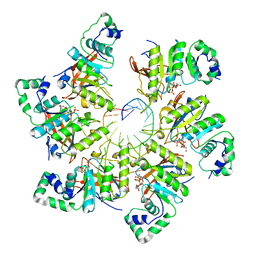 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (form III) | | Descriptor: | DNA (25-MER), DNA primase/helicase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
8YJM
 
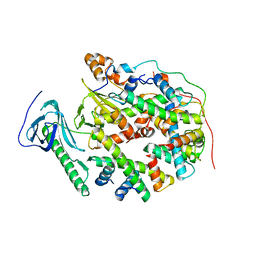 | | Structure of human SPT16 MD-CTD and MCM2 HBD chaperoning a histone H3-H4 tetramer and a single chain H2B-H2A chimera | | Descriptor: | DNA replication licensing factor MCM2, FACT complex subunit SPT16, Histone H2B 1/2/3/4/6,Histone H2A type 1-D, ... | | Authors: | Gan, S.L, Yang, W.S, Xu, R.M. | | Deposit date: | 2024-03-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structure of a histone hexamer bound by the chaperone domains of SPT16 and MCM2.
Sci China Life Sci, 67, 2024
|
|
8YJF
 
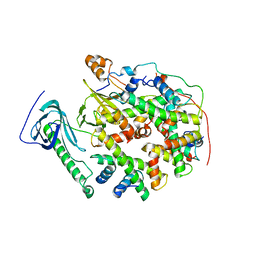 | | Structure of human SPT16 MD-CTD and MCM2 HBD chaperoning a histone H3-H4 tetramer and an H2A-H2B dimer | | Descriptor: | DNA replication licensing factor MCM2, FACT complex subunit SPT16, Histone H2A type 1-D, ... | | Authors: | Gan, S.L, Yang, W.S, Xu, R.M. | | Deposit date: | 2024-03-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure of a histone hexamer bound by the chaperone domains of SPT16 and MCM2.
Sci China Life Sci, 67, 2024
|
|
2QK9
 
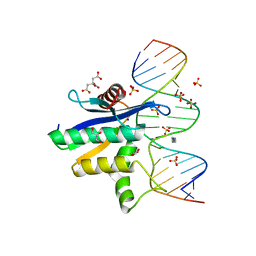 | | Human RNase H catalytic domain mutant D210N in complex with 18-mer RNA/DNA hybrid | | Descriptor: | 5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*GP*CP*AP*CP*T)-3', 5'-R(*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3', CITRATE ANION, ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Ghirlando, R, Cerritelli, S.M, Crouch, R.J, Yang, W. | | Deposit date: | 2007-07-10 | | Release date: | 2007-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription
Mol.Cell, 28, 2007
|
|
2QKB
 
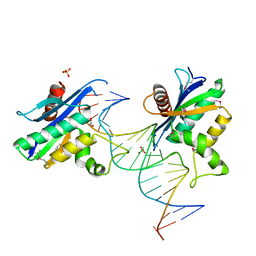 | | Human RNase H catalytic domain mutant D210N in complex with 20-mer RNA/DNA hybrid | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*DGP*DGP*DAP*DAP*DTP*DCP*DAP*DGP*DGP*DTP*DGP*DTP*DCP*DGP*DCP*DAP*DCP*DTP*DCP*DT)-3', 5'-R(*GP*GP*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3'), ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Ghirlando, R, Cerritelli, S.M, Crouch, R.J, Yang, W. | | Deposit date: | 2007-07-10 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription
Mol.Cell, 28, 2007
|
|
2YOJ
 
 | | HCV NS5B polymerase complexed with pyridonylindole compound | | Descriptor: | 4-fluoranyl-6-[(7-fluoranyl-4-oxidanylidene-3H-quinazolin-6-yl)methyl]-8-(2-oxidanylidene-1H-pyridin-3-yl)furo[2,3-e]indole-7-carboxylic acid, PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Chen, K.X, Venkatraman, S, Anilkumar, G.N, Zeng, Q, Lesburg, C.A, Vibulbhan, B, Yang, W, Velazquez, F, Chan, T.-Y, Bennett, F, Sannigrahi, M, Jiang, Y, Duca, J.S, Pinto, P, Gavalas, S, Huang, Y, Wu, W, Selyutin, O, Agrawal, S, Feld, B, Huang, H.-C, Li, C, Cheng, K.-C, Shih, N.-Y, Kozlowski, J.A, Rosenblum, S.B, Njoroge, F.G. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of Sch 900188: A Potent Hepatitis C Virus Ns5B Polymerase Inhibitor Prodrug as a Development Candidate
Acs Med.Chem.Lett., 5, 2014
|
|
7K1B
 
 | | CryoEM structure of DNA-PK catalytic subunit complexed with DNA (Complex II) | | Descriptor: | DNA (5'-D(P*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K11
 
 | |
7K0Y
 
 | | Cryo-EM structure of activated-form DNA-PK (complex VI) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-06 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K10
 
 | |
7K19
 
 | | CryoEM structure of DNA-PK catalytic subunit complexed with DNA (Complex I) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K1J
 
 | | CryoEM structure of inactivated-form DNA-PK (Complex III) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K1N
 
 | | CryoEM structure of inactivated-form DNA-PK (Complex V) | | Descriptor: | DNA (5'-D(P*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-08 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K17
 
 | |
7K1K
 
 | | CryoEM structure of inactivated-form DNA-PK (Complex IV) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
6IQT
 
 | | Crystal Structure of CagV, a VirB8 homolog of T4SS from Helicobacter pylori Strain 26695 | | Descriptor: | Cag pathogenicity island protein (Cag10) | | Authors: | Wu, X, Zhao, Y, Sun, L, Jiang, M, Wang, Q, Wang, Q, Yang, W, Wu, Y. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-13 | | Last modified: | 2020-04-29 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Crystal structure of CagV, the Helicobacter pylori homologue of the T4SS protein VirB8.
Febs J., 286, 2019
|
|
2YPS
 
 | | Crystal structure of the PX domain of human sorting nexin 3 | | Descriptor: | SORTING NEXIN-3 | | Authors: | Canning, P, Kiyani, W, Froese, D.S, Krojer, T, Strain-Damerell, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Yue, W.W. | | Deposit date: | 2012-10-31 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Px Domain of Human Sorting Nexin 3
To be Published
|
|
6BSG
 
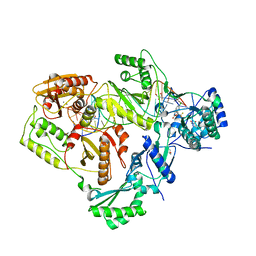 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in an RNA hydrolysis-off mode | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSH
 
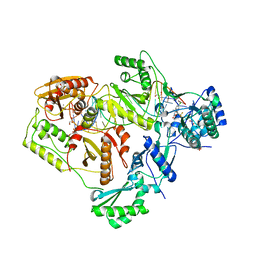 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in the RNA hydrolysis mode | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*AP*TP*GP*CP*CP*AP*CP*TP*AP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSJ
 
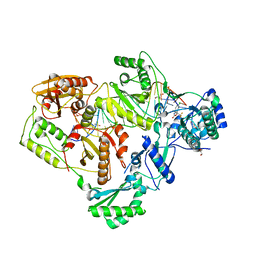 | | Structure of HIV-1 RT complexed with an RNA/DNA hybrid sequence non-preferred for RNA hydrolysis | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSI
 
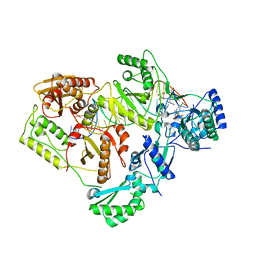 | | Structure of HIV-1 RT complexed with an RNA/DNA hybrid containing the polypurine-tract sequence | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*TP*TP*TP*TP*CP*TP*TP*TP*TP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2CNQ
 
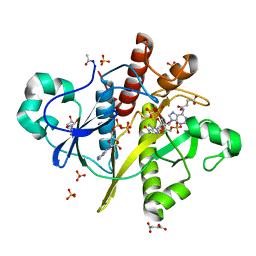 | | Atomic resolution structure of SAICAR-synthase from Saccharomyces cerevisiae complexed with ADP, AICAR, succinate | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, W.R. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Saicar Synthase: Substrate Recognition, Conformational Flexibility and Catalysis.
To be Published
|
|
