8IMI
 
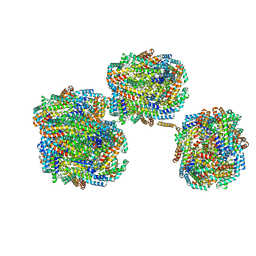 | | A1-A2, A3-A4, B'1-B'2, C'1-C'2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster A) | | Descriptor: | ApcA2, ApcB2, ApcB3, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
7VJ6
 
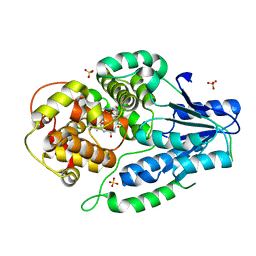 | | SFX structure of archaeal class II CPD photolyase from Methanosarcina mazei in the semiquinone state | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ8
 
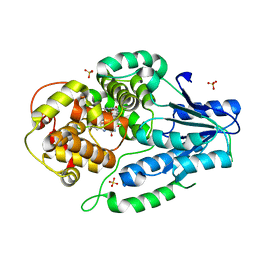 | | SFX structure of archaeal class II CPD photolyase from Methanosarcina mazei in the oxidized state | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ7
 
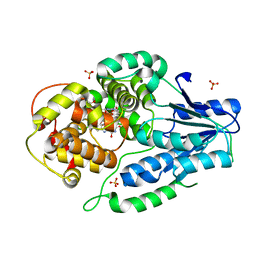 | | SFX structure of archaeal class II CPD photolyase from Methanosarcina mazei in the fully reduced state | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
2VUH
 
 | |
7WQA
 
 | | SARS-CoV-2 main protease in complex with Z-VAD-FMK | | Descriptor: | 3C-like proteinase, Z-VAD(OMe)-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 main protease in complex with Z-VAD-FMK
To Be Published
|
|
7WQ9
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with Z-IETD-FMK | | Descriptor: | 3C-like proteinase, Z-IETD-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with Z-IETD-FMK
To Be Published
|
|
7WQ8
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with Z-DEVD-FMK | | Descriptor: | 3C-like proteinase, Z-DEVD-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with Z-DEVD-FMK
To Be Published
|
|
7WQB
 
 | | SARS-CoV-2 main protease mutant (P168A) in complex with MG-132 | | Descriptor: | 3C-like proteinase nsp5, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | SARS-CoV-2 main protease mutant (P168A) in complex with MG-132
To Be Published
|
|
3BIE
 
 | |
4MG2
 
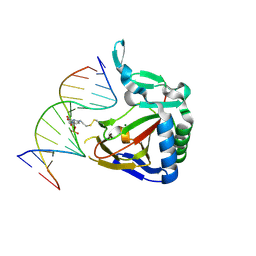 | | ALKBH2 F102A cross-linked to undamaged dsDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA-1, DNA-2, ... | | Authors: | Chen, B, Gan, J, Yang, C.G. | | Deposit date: | 2013-08-28 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The complex structures of ALKBH2 mutants cross-linked to
dsDNA reveal the conformational swing of β-hairpin
Sci China Chem, 57, 2014
|
|
7CAM
 
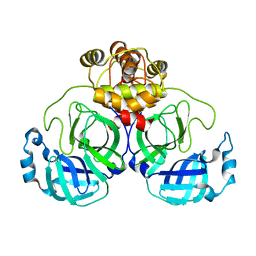 | | SARS-CoV-2 main protease (Mpro) apo structure (space group P212121) | | Descriptor: | 3C-like proteinase | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y. | | Deposit date: | 2020-06-09 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
7CB7
 
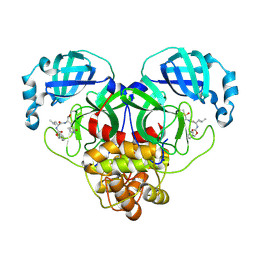 | | 1.7A resolution structure of SARS-CoV-2 main protease (Mpro) in complex with broad-spectrum coronavirus protease inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y, Hung, M.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
7DDC
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with Tafenoquine | | Descriptor: | 3C-like proteinase, Tafenoquine | | Authors: | Chen, Y, Wang, Y.C, Yang, C.S, Hung, M.C. | | Deposit date: | 2020-10-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Tafenoquine and its derivatives as inhibitors for the severe acute respiratory syndrome coronavirus 2.
J.Biol.Chem., 298, 2022
|
|
6KKO
 
 | |
7F8T
 
 | | Re-refinement of the 2XRY X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-07-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
4JXI
 
 | | Directed evolution and rational design of a de novo designed esterase toward improved catalysis. Northeast Structural Genomics Consortium (NESG) Target OR184 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, BROMIDE ION, GLYCEROL, ... | | Authors: | Kuzin, A, Smith, M.D, Richter, F, Lew, S, Seetharaman, R, Bryan, C, Lech, Z, Kiss, G, Moretti, R, Maglaqui, M, Xiao, R, Kohan, E, Smith, M, Everett, J.K, Nguyen, R, Pande, V, Hilvert, D, Kornhaber, G, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Directed evolution and rational design of a de novo designed esterase toward improved catalysis. Northeast Structural Genomics Consortium (NESG) Target OR184
To be Published
|
|
7DD0
 
 | |
2VUI
 
 | | Crystal structure of the HupR receiver domain in inhibitory phospho- state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, HYDROGENASE TRANSCRIPTIONAL REGULATORY PROTEIN HUPR1, MAGNESIUM ION | | Authors: | Davies, K.M, Lowe, E.D, Venien-Bryan, C, Johnson, L.N. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Hupr Receiver Domain Crystal Structure in its Nonphospho and Inhibitory Phospho States.
J.Mol.Biol., 385, 2009
|
|
7EBL
 
 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), STING | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-10 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
7EBD
 
 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ACETATE ION, TIR-like domain-containing protein | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
8EG6
 
 | | huCaspase-6 in complex with inhibitor 2a | | Descriptor: | (3R)-1-(ethanesulfonyl)-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | Deposit date: | 2022-09-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
8EG5
 
 | | huCaspase-6 in complex with inhibitor 3a | | Descriptor: | (3R,5S)-1-(ethanesulfonyl)-5-phenyl-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide (bound form), 1,2-ETHANEDIOL, Caspase-6 subunit p11, ... | | Authors: | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | Deposit date: | 2022-09-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6V67
 
 | | Apo Structure of the De Novo PD-1 Binding Miniprotein GR918.2 | | Descriptor: | PD-1 Binding Miniprotein GR918.2 | | Authors: | Bick, M.J, Bryan, C.M, Baker, D, Dimaio, F, Kang, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Computational design of a synthetic PD-1 agonist.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
