6THZ
 
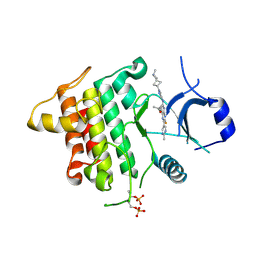 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 7-fluoranyl-~{N}-[1-(2-methyl-2-azaspiro[3.3]heptan-6-yl)pyrazol-4-yl]-4-(1-methylcyclopropyl)oxy-6-(2-methylpyrimidin-5-yl)pyrido[3,2-d]pyrimidin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Degorce, S.L. | | Deposit date: | 2019-11-21 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Improving metabolic stability and removing aldehyde oxidase liability in a 5-azaquinazoline series of IRAK4 inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
7QG3
 
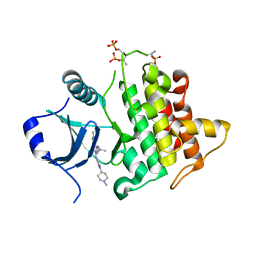 | | IRAK4 in complex with inhibitor | | Descriptor: | 6-[(2~{S})-2-fluoranylpropyl]-4-[(1-methylcyclopropyl)amino]-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]pyrido[4,3-d]pyrimidin-5-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
7QG1
 
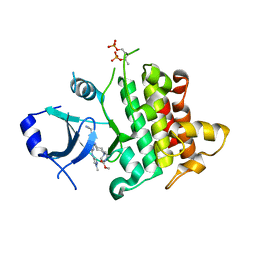 | | IRAK4 in complex with inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, methyl 4-[4-[[6-(cyanomethyl)-2-[(1-methylpyrazol-4-yl)amino]-5-oxidanylidene-pyrido[4,3-d]pyrimidin-4-yl]amino]cyclohexyl]piperazine-1-carboxylate | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
6THW
 
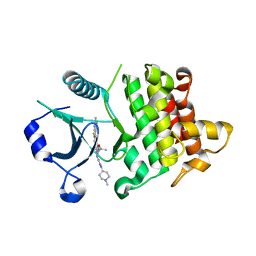 | | IRAK4 in complex with inhibitor | | Descriptor: | 7-fluoranyl-4-(1-methylcyclopropyl)oxy-~{N}-[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]-6-(2-methylpyrimidin-5-yl)pyrido[3,2-d]pyrimidin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Degorce, S.L. | | Deposit date: | 2019-11-21 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Improving metabolic stability and removing aldehyde oxidase liability in a 5-azaquinazoline series of IRAK4 inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
6F3G
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}4,~{N}4-dimethyl-~{N}1-(5-propan-2-ylpyrrolo[3,2-d]pyrimidin-4-yl)cyclohexane-1,4-diamine | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
6F3D
 
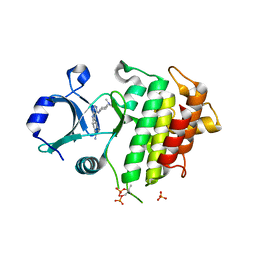 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 4-[4-[[4-(dimethylamino)cyclohexyl]amino]-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]cyclohexane-1-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
6F3I
 
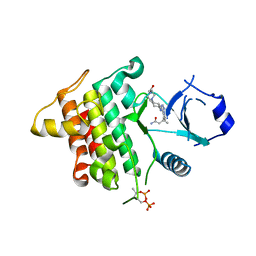 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | (3~{R})-3-[4-[[4-(4-ethanoylpiperazin-1-yl)cyclohexyl]amino]pyrrolo[2,1-f][1,2,4]triazin-5-yl]butanamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
6F3E
 
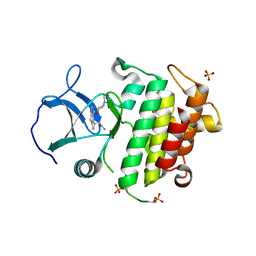 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 2-[(3~{R})-12-(4-morpholin-4-ylcyclohexyl)oxy-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9,11-tetraen-3-yl]ethanamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
4H1D
 
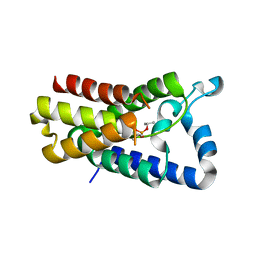 | | Cocrystal structure of GlpG and DFP | | Descriptor: | DIISOPROPYL PHOSPHONATE, Rhomboid protease GlpG | | Authors: | Xue, Y, Ha, Y. | | Deposit date: | 2012-09-10 | | Release date: | 2013-05-01 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.8975 Å) | | Cite: | Large lateral movement of transmembrane helix s5 is not required for substrate access to the active site of rhomboid intramembrane protease.
J.Biol.Chem., 288, 2013
|
|
1CSM
 
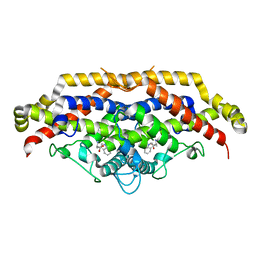 | |
4BTK
 
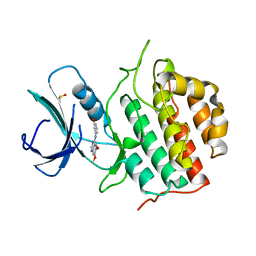 | | TTBK1 in complex with inhibitor | | Descriptor: | 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE, DIMETHYL SULFOXIDE, TAU-TUBULIN KINASE 1 | | Authors: | Xue, Y, Wan, P, Hillertz, P, Schweikart, F, Zhao, Y, Wissler, L, Dekker, N. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structural Analysis of Tau-Tubulin Kinase 1 and its Interactions with Small Molecular Inhibitors.
Chemmedchem, 8, 2013
|
|
4BTJ
 
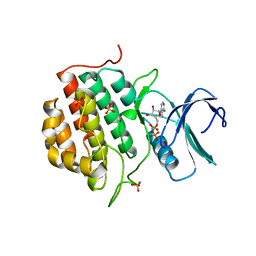 | | TTBK1 in complex with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Xue, Y, Wan, P, Hillertz, P, Schweikart, F, Zhao, Y, Wissler, L, Dekker, N. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | X-Ray Structural Analysis of Tau-Tubulin Kinase 1 and its Interactions with Small Molecular Inhibitors.
Chemmedchem, 8, 2013
|
|
4BTM
 
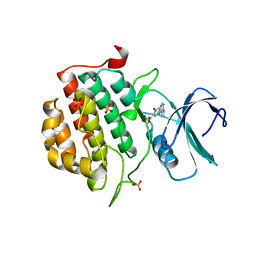 | | TTBK1 in complex with inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Xue, Y, Wan, P, Hillertz, P, Schweikart, F, Zhao, Y, Wissler, L, Dekker, N. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | X-Ray Structural Analysis of Tau-Tubulin Kinase 1 and its Interactions with Small Molecular Inhibitors.
Chemmedchem, 8, 2013
|
|
3BG8
 
 | | Crystal structure of Factor XIa in complex with Clavatadine A | | Descriptor: | BENZAMIDINE, Coagulation factor XIa light chain, N-(4-carbamimidamidobutyl)ethanamide, ... | | Authors: | Xue, Y, Oster, L. | | Deposit date: | 2007-11-26 | | Release date: | 2008-12-09 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Clavatadine A, a natural product with selective recognition and irreversible inhibition of factor XIa.
J.Med.Chem., 51, 2008
|
|
3Q7G
 
 | | Crystal Structure of E2 domain of Human Amyloid Precursor-Like Protein 1 in complex with SOS (sucrose octasulfate) | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Amyloid-like protein 1 | | Authors: | Xue, Y, Ha, Y. | | Deposit date: | 2011-01-04 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the E2 Domain of Amyloid Precursor Protein-like Protein 1 in Complex with Sucrose Octasulfate.
J.Biol.Chem., 286, 2011
|
|
3Q7L
 
 | |
3UBB
 
 | |
2AFR
 
 | |
2AFV
 
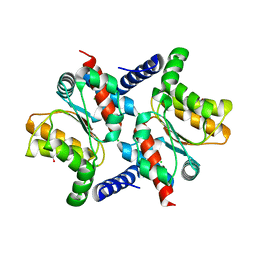 | | The Crystal Structure of Putative Precorrin Isomerase CbiC in Cobalamin Biosynthesis | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Xue, Y, Wei, Z. | | Deposit date: | 2005-07-26 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of putative precorrin isomerase CbiC in cobalamin biosynthesis
J.Struct.Biol., 153, 2006
|
|
3QMK
 
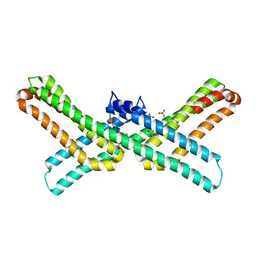 | | Crystal structure of the E2 domain of APLP1 in complex with heparin hexasaccharide | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Amyloid-like protein 1, SULFATE ION | | Authors: | Xue, Y, Ha, Y. | | Deposit date: | 2011-02-04 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of amyloid precursor-like protein 1 and heparin complex suggests a dual role of heparin in E2 dimerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1BIC
 
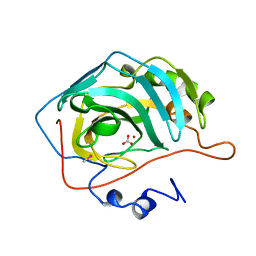 | | CRYSTALLOGRAPHIC ANALYSIS OF THR-200-> HIS HUMAN CARBONIC ANHYDRASE II AND ITS COMPLEX WITH THE SUBSTRATE, HCO3- | | Descriptor: | BICARBONATE ION, CARBONIC ANHYDRASE II, METHYL MERCURY ION, ... | | Authors: | Xue, Y, Vidgren, J, Svensson, L.A, Liljas, A, Jonsson, B.-H, Lindskog, S. | | Deposit date: | 1992-09-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis of Thr-200-->His human carbonic anhydrase II and its complex with the substrate, HCO3-.
Proteins, 15, 1993
|
|
1CAK
 
 | | STRUCTURAL ANALYSIS OF THE ZINC HYDROXIDE-THR 199-GLU 106 HYDROGEN BONDING NETWORK IN HUMAN CARBONIC ANHYDRASE II | | Descriptor: | CARBONIC ANHYDRASE II, SULFATE ION, ZINC ION | | Authors: | Xue, Y, Liljas, A, Jonsson, B.-H, Lindskog, S. | | Deposit date: | 1992-09-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the zinc hydroxide-Thr-199-Glu-106 hydrogen-bond network in human carbonic anhydrase II.
Proteins, 17, 1993
|
|
1CAJ
 
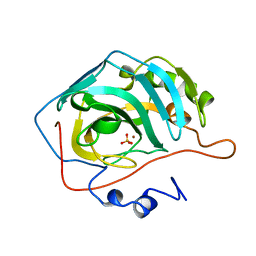 | | STRUCTURAL ANALYSIS OF THE ZINC HYDROXIDE-THR 199-GLU 106 HYDROGEN BONDING NETWORK IN HUMAN CARBONIC ANHYDRASE II | | Descriptor: | CARBONIC ANHYDRASE II, SULFATE ION, ZINC ION | | Authors: | Xue, Y, Liljas, A, Jonsson, B.-H, Lindskog, S. | | Deposit date: | 1992-09-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the zinc hydroxide-Thr-199-Glu-106 hydrogen-bond network in human carbonic anhydrase II.
Proteins, 17, 1993
|
|
1CAM
 
 | | STRUCTURAL ANALYSIS OF THE ZINC HYDROXIDE-THR 199-GLU 106 HYDROGEN BONDING NETWORK IN HUMAN CARBONIC ANHYDRASE II | | Descriptor: | BICARBONATE ION, CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Xue, Y, Liljas, A, Jonsson, B.-H, Lindskog, S. | | Deposit date: | 1992-09-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of the zinc hydroxide-Thr-199-Glu-106 hydrogen-bond network in human carbonic anhydrase II.
Proteins, 17, 1993
|
|
1CAL
 
 | | STRUCTURAL ANALYSIS OF THE ZINC HYDROXIDE-THR 199-GLU 106 HYDROGEN BONDING NETWORK IN HUMAN CARBONIC ANHYDRASE II | | Descriptor: | CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Xue, Y, Liljas, A, Jonsson, B.-H, Lindskog, S. | | Deposit date: | 1992-09-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the zinc hydroxide-Thr-199-Glu-106 hydrogen-bond network in human carbonic anhydrase II.
Proteins, 17, 1993
|
|
