1NW1
 
 | | Crystal Structure of Choline Kinase | | Descriptor: | CALCIUM ION, Choline kinase (49.2 kD) | | Authors: | Peisach, D, Gee, P, Kent, C, Xu, Z. | | Deposit date: | 2003-02-05 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Crystal Structure of Choline Kinase Reveals a Eukaryotic Protein Kinase Fold
Structure, 11, 2003
|
|
7YSF
 
 | | Crystal structure of ZNF524 ZF1-4 in complex with telomeric DNA | | Descriptor: | D-MALATE, DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*C)-3'), ... | | Authors: | Li, F.D, Xu, Z.Y. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-16 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ZNF524 directly interacts with telomeric DNA and supports telomere integrity.
Nat Commun, 14, 2023
|
|
4U7E
 
 | | The crystal structure of the complex of LIP5 NTD and IST1 MIM | | Descriptor: | IST1 homolog, Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Guo, E.Z, Xu, Z. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinct Mechanisms of Recognizing Endosomal Sorting Complex Required for Transport III (ESCRT-III) Protein IST1 by Different Microtubule Interacting and Trafficking (MIT) Domains.
J.Biol.Chem., 290, 2015
|
|
4U7I
 
 | | Structure of the complex of Spartin MIT and IST1 MIM | | Descriptor: | IST1 homolog, Spartin | | Authors: | Guo, E.Z, Xu, Z. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Distinct Mechanisms of Recognizing Endosomal Sorting Complex Required for Transport III (ESCRT-III) Protein IST1 by Different Microtubule Interacting and Trafficking (MIT) Domains.
J.Biol.Chem., 290, 2015
|
|
4U7Y
 
 | | Structure of the complex of VPS4B MIT and IST1 MIM | | Descriptor: | IST1 homolog, Vacuolar protein sorting-associated protein 4B | | Authors: | Guo, E.Z, Xu, Z. | | Deposit date: | 2014-07-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Distinct Mechanisms of Recognizing Endosomal Sorting Complex Required for Transport III (ESCRT-III) Protein IST1 by Different Microtubule Interacting and Trafficking (MIT) Domains.
J.Biol.Chem., 290, 2015
|
|
1U3R
 
 | | Crystal Structure of Estrogen Receptor beta complexed with WAY-338 | | Descriptor: | 2-(5-HYDROXY-NAPHTHALEN-1-YL)-1,3-BENZOOXAZOL-6-OL, Estrogen receptor beta, steroid receptor coactivator-1 | | Authors: | Malamas, M.S, Manas, E.S, McDevitt, R.E, Gunawan, I, Xu, Z.B, Collini, M.D, Miller, C.P, Dinh, T, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Design and synthesis of aryl diphenolic azoles as potent and selective estrogen receptor-beta ligands.
J.Med.Chem., 47, 2004
|
|
1U9E
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-397 | | Descriptor: | 2-(4-HYDROXY-PHENYL)BENZOFURAN-5-OL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-09 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design Of Estrogen Receptor-beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1U3S
 
 | | Crystal Structure of Estrogen Receptor beta complexed with WAY-797 | | Descriptor: | 3-(6-HYDROXY-NAPHTHALEN-2-YL)-BENZO[D]ISOOXAZOL-6-OL, Estrogen receptor beta, steroid receptor coactivator-1 | | Authors: | Malamas, M.S, Manas, E.S, McDevitt, R.E, Gunawan, I, Xu, Z.B, Collini, M.D, Miller, C.P, Dinh, T, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and synthesis of aryl diphenolic azoles as potent and selective estrogen receptor-beta ligands.
J.Med.Chem., 47, 2004
|
|
1UKF
 
 | | Crystal Structure of Pseudomonas Avirulence Protein AvrPphB | | Descriptor: | Avirulence protein AVRPPH3 | | Authors: | Zhu, M, Shao, F, Innes, R.W, Dixon, J.E, Xu, Z. | | Deposit date: | 2003-08-21 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The crystal structure of Pseudomonas avirulence protein AvrPphB: a papain-like fold with a distinct substrate-binding site.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1R62
 
 | | Crystal structure of the C-terminal Domain of the Two-Component System Transmitter Protein NRII (NtrB) | | Descriptor: | Nitrogen regulation protein NR(II) | | Authors: | Song, Y, Peisach, D, Pioszak, A.A, Xu, Z, Ninfa, A.J. | | Deposit date: | 2003-10-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the C-terminal Domain of the Two-Component System Transmitter Protein Nitrogen Regulator II (NRII; NtrB), Regulator of Nitrogen Assimilation in Escherichia coli.
Biochemistry, 43, 2004
|
|
3GGY
 
 | | Crystal Structure of S.cerevisiae Ist1 N-terminal domain | | Descriptor: | Increased sodium tolerance protein 1 | | Authors: | Xiao, J, Xu, Z. | | Deposit date: | 2009-03-02 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis.
MOLECULAR BIOLOGY OF THE CELL, 20, 2009
|
|
3GGZ
 
 | |
1U3Q
 
 | | Crystal Structure of Estrogen Receptor beta complexed with CL-272 | | Descriptor: | 4-(6-HYDROXY-BENZO[D]ISOXAZOL-3-YL)BENZENE-1,3-DIOL, Estrogen receptor beta | | Authors: | Malamas, M.S, Manas, E.S, McDevitt, R.E, Gunawan, I, Xu, Z.B, Collini, M.D, Miller, C.P, Dinh, T, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of aryl diphenolic azoles as potent and selective estrogen receptor-beta ligands.
J.Med.Chem., 47, 2004
|
|
8KG9
 
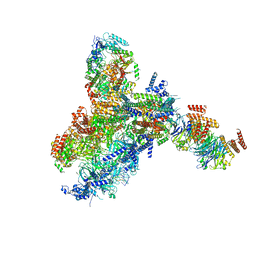 | | Yeast replisome in state III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
3VPM
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human ribonucleotide reductase M2 subunit
To be Published
|
|
3VPO
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
3VPN
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
8KG6
 
 | | Yeast replisome in state I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
4NIQ
 
 | | Crystal Structure of Vps4 MIT-Vfa1 MIM2 | | Descriptor: | VPS4-associated protein 1, Vacuolar protein sorting-associated protein 4 | | Authors: | Vild, C.J, Xu, Z. | | Deposit date: | 2013-11-06 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Vfa1 Binds to the N-terminal Microtubule-interacting and Trafficking (MIT) Domain of Vps4 and Stimulates Its ATPase Activity.
J.Biol.Chem., 289, 2014
|
|
4YFJ
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib | | Descriptor: | Aminoglycoside 3'-N-acetyltransferase, SULFATE ION | | Authors: | Stogios, P.J, Xu, Z, Evdokimova, E, Yim, V, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib
To Be Published
|
|
7KES
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in complex with apramycin and CoA | | Descriptor: | APRAMYCIN, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-12 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
8HWS
 
 | | The complex structure of Omicron BA.4 RBD with BD604, S309, and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab Heavy chain, BD-604 Fab Light chain, ... | | Authors: | He, Q.W, Xu, Z.P, Xie, Y.F. | | Deposit date: | 2023-01-02 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.
Cell Rep Med, 4, 2023
|
|
4GX9
 
 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Li, N, Horan, N, Xu, Z.-Q, Jacques, D, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
3DYR
 
 | |
