7XM7
 
 | | Crystal Structure of the CBP in complex with the Y08188 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethanoyl-~{N}-[2-fluoranyl-3-(1-methylpyrazol-4-yl)phenyl]-7-methoxy-indolizine-1-carboxamide, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as potent and selective CBP bromodomain inhibitors
To Be Published
|
|
7XNE
 
 | | Crystal structure of CBP bromodomain liganded with Y08284 | | Descriptor: | CREB-binding protein, GLYCEROL, N-[3-(1-cyclopropylpyrazol-4-yl)-2-fluoranyl-5-[(1S)-1-oxidanylethyl]phenyl]-3-ethanoyl-7-methoxy-indolizine-1-carboxamide | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, C, Hu, Q, Luo, G, Hu, J, Zhuang, X, Zou, L, Shen, H, Wu, X, Zhang, Y, Kong, X, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
5ILI
 
 | | Tobacco 5-epi-aristolochene synthase with CAPSO buffer molecule and Mg2+ ions | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 5-epi-aristolochene synthase, ... | | Authors: | Koo, H.J, Louie, G.V, Xu, Y, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small-molecule buffer components can directly affect terpene-synthase activity by interacting with the substrate-binding site of the enzyme
To Be Published
|
|
5ILD
 
 | | Tobacco 5-epi-aristolochene synthase with MES buffer molecule and Mg2+ ions | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-epi-aristolochene synthase, MAGNESIUM ION | | Authors: | Koo, H.J, Louie, G.V, Xu, Y, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Small-molecule buffer components can directly affect terpene-synthase activity by interacting with the substrate-binding site of the enzyme
To Be Published
|
|
5IM1
 
 | |
2MC5
 
 | | A bacteriophage transcription regulator inhibits bacterial transcription initiation by -factor displacement | | Descriptor: | 45L | | Authors: | Liu, B, Shadrin, A, Sheppard, C, Xu, Y, Severinov, K, Matthews, S, Wigneshweraraj, S. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A bacteriophage transcription regulator inhibits bacterial transcription initiation by sigma-factor displacement.
Nucleic Acids Res., 42, 2014
|
|
4QG9
 
 | | crystal structure of PKM2-R399E mutant | | Descriptor: | ACETATE ION, MAGNESIUM ION, Pyruvate kinase PKM | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
4NDZ
 
 | | Structure of Maltose Binding Protein fusion to 2-O-Sulfotransferase with bound heptasaccharide and PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1 fusion, ... | | Authors: | Liu, C, Sheng, J, Krahn, J.M, Perera, L, Xu, Y, Hsieh, P, Liu, J, Pedersen, L.C. | | Deposit date: | 2013-10-28 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Deciphering the role of 2-O-sulfotransferase in regulating heparan sulfate biosynthesis
To be Published
|
|
2LLY
 
 | | NMR structures of the transmembrane domains of the nAChR a4 subunit | | Descriptor: | Neuronal acetylcholine receptor subunit alpha-4 | | Authors: | Bondarenko, V, Mowrey, D, Tillman, T, Cui, T, Liu, L.T, Xu, Y, Tang, P. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structures of the transmembrane domains of the a4b2 nAChR.
Biochim.Biophys.Acta, 1818, 2012
|
|
2KZ1
 
 | | Inter-molecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric back-protonation and 2D NOESY | | Descriptor: | Interferon alpha-2, Soluble IFN alpha/beta receptor | | Authors: | Nudelman, I, Akabayov, S.R, Schnur, E, Biron, Z, Levy, R, Xu, Y, Yang, D, Anglister, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Intermolecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric reverse-protonation and two-dimensional NOESY
Biochemistry, 49, 2010
|
|
2KSR
 
 | |
2MAW
 
 | |
6ICX
 
 | |
6ID9
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SGTL ( A138S, V186G, P221T) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6ICW
 
 | |
6ID2
 
 | |
8HDU
 
 | |
8HDV
 
 | |
4F8H
 
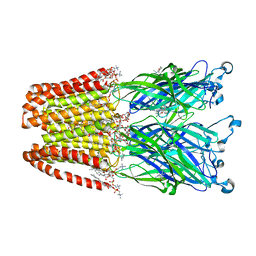 | | X-ray Structure of the Anesthetic Ketamine Bound to the GLIC Pentameric Ligand-gated Ion Channel | | Descriptor: | (R)-ketamine, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Proton-gated ion channel, ... | | Authors: | Pan, J.J, Chen, Q, Willenbring, D, Kong, X.P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2012-05-17 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the Pentameric Ligand-Gated Ion Channel GLIC Bound with Anesthetic Ketamine.
Structure, 20, 2012
|
|
4DUG
 
 | | Crystal Structure of Circadian Clock Protein KaiC E318A Mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION, ... | | Authors: | Egli, M, Mori, T, Pattanayek, R, Xu, Y, Qin, X, Johnson, C.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.292 Å) | | Cite: | Dephosphorylation of the Core Clock Protein KaiC in the Cyanobacterial KaiABC Circadian Oscillator Proceeds via an ATP Synthase Mechanism.
Biochemistry, 51, 2012
|
|
6ICY
 
 | | Crystal structure of H7 hemagglutinin mutant H7-AGTL ( V186G, P221T) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-07 | | Release date: | 2019-11-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6IDB
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTQ ( A138S, P221T, L226Q) with 6'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6IDZ
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTQ ( A138S, P221T, L226Q) with 3'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6ID5
 
 | |
6IDA
 
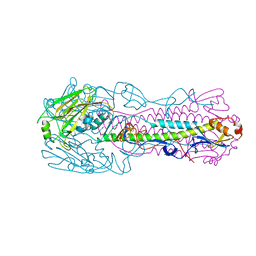 | | Crystal structure of H7 hemagglutinin mutant H7-SVTQ ( A138S, P221T, L226Q) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
