1HMP
 
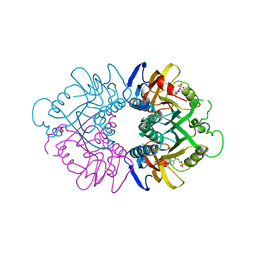 | | THE CRYSTAL STRUCTURE OF HUMAN HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE WITH BOUND GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE GUANINE PHOSPHORIBOSYL-TRANSFERASE | | Authors: | Eads, J.C, Scapin, G, Xu, Y, Grubmeyer, C, Sacchettini, J.C. | | Deposit date: | 1994-06-03 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of human hypoxanthine-guanine phosphoribosyltransferase with bound GMP.
Cell(Cambridge,Mass.), 78, 1994
|
|
8Y53
 
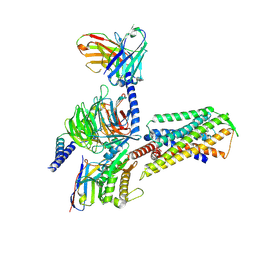 | | Cryo-EM structure of the MK-5046-bound BRS3-Gq complex | | Descriptor: | (2~{S})-1,1,1-tris(fluoranyl)-2-(4-pyrazol-1-ylphenyl)-3-[5-[[1-(trifluoromethyl)cyclopropyl]methyl]-1~{H}-imidazol-2-yl]propan-2-ol, Bombesin receptor subtype-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Xu, Y, Yin, W, Xu, H.E. | | Deposit date: | 2024-01-31 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into ligand recognition, selectivity, and activation of bombesin receptor subtype-3.
Cell Rep, 43, 2024
|
|
8Y52
 
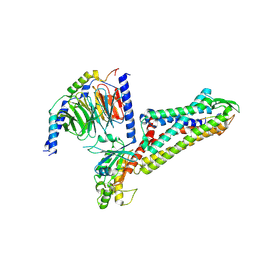 | | Cryo-EM structure of the BA1-bound BRS3-Gq complex | | Descriptor: | BA1, Bombesin receptor subtype-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Xu, Y, Yin, W, Xu, H.E. | | Deposit date: | 2024-01-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into ligand recognition, selectivity, and activation of bombesin receptor subtype-3.
Cell Rep, 43, 2024
|
|
4MXC
 
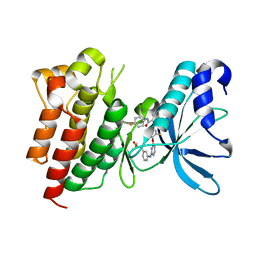 | | Crystal structure of CMET in complex with novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[2-({3-[(methylsulfonyl)methyl]phenyl}amino)pyrimidin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2013-09-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Discovery of Anilinopyrimidines as Dual Inhibitors of c-Met and VEGFR-2: Synthesis, SAR, and Cellular Activity
ACS MED.CHEM.LETT., 5, 2014
|
|
7XHE
 
 | | Crystal structure of CBP bromodomain liganded with CCS151 | | Descriptor: | (6S)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]-1-(3-fluoranyl-4-methoxy-phenyl)piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-08 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XH6
 
 | | Crystal structure of CBP bromodomain liganded with CCS1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XI0
 
 | | Crystal structure of CBP bromodomain liganded with CCS150 | | Descriptor: | (6S)-1-(3-chloranyl-4-methoxy-phenyl)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, GLYCEROL | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-11 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
6M08
 
 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor. | | Descriptor: | (2S)-2-[(Z)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
6M07
 
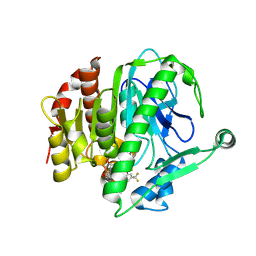 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | Descriptor: | (2S)-2-[(E)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)-2-[2,2,2-tris(fluoranyl)ethoxy]phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
2A7P
 
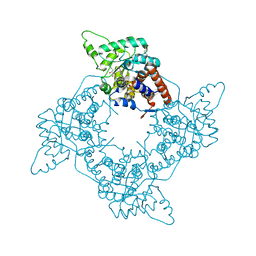 | | Crystal Structure of the G81A mutant of the Active Chimera of (S)-Mandelate Dehydrogenase in complex with its substrate 3-indolelactate | | Descriptor: | (S)-Mandelate Dehydrogenase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(INDOL-3-YL) LACTATE, ... | | Authors: | Sukumar, N, Xu, Y, Mitra, B, Mathews, F.S. | | Deposit date: | 2005-07-05 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the G81A mutant form of the active chimera of (S)-mandelate dehydrogenase and its complex with two of its substrates.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
8J0I
 
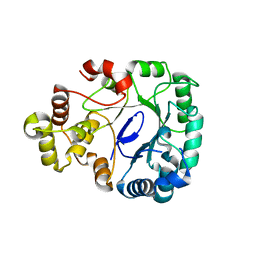 | | Aldo-keto reductase KmAKR | | Descriptor: | NADPH-dependent alpha-keto amide reductase, SODIUM ION | | Authors: | Xu, S.Y, Zhou, L, Xu, Y, Wang, Y.J, Zheng, Y.G. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Aldo-keto reductase KmAKR from Kluyveromyces marxianus
To Be Published
|
|
6HN5
 
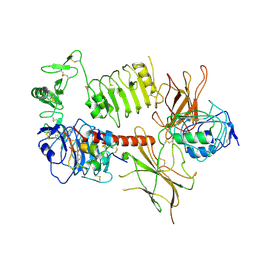 | | Leucine-zippered human insulin receptor ectodomain with single bound insulin - "upper" membrane-distal part | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, ... | | Authors: | Weis, F, Menting, J.G, Margetts, M.B, Chan, S.J, Xu, Y, Tennagels, N, Wohlfart, P, Langer, T, Mueller, C.W, Dreyer, M.K, Lawrence, M.C. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The signalling conformation of the insulin receptor ectodomain.
Nat Commun, 9, 2018
|
|
6LRC
 
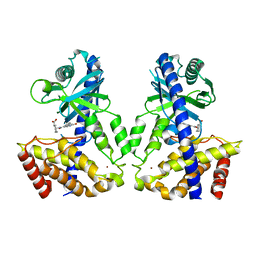 | | Human cGAS catalytic domain bound with the inhibitor PF-06928215 | | Descriptor: | (1R,2S)-2-[(7-hydroxy-5-phenylpyrazolo[1,5-a]pyrimidine-3-carbonyl)amino]cyclohexane-1-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xu, Y.C. | | Deposit date: | 2020-01-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
8YHF
 
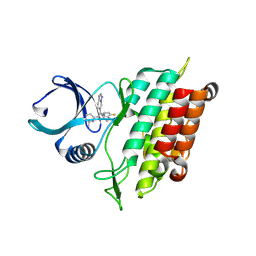 | | The Crystal Structure of the Type I TGF beta receptor from Biortus. | | Descriptor: | 2-fluoranyl-~{N}-[[5-(6-methylpyridin-2-yl)-4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-1~{H}-imidazol-2-yl]methyl]aniline, TGF-beta receptor type-1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Type I TGF beta receptor from Biortus.
To Be Published
|
|
8YGY
 
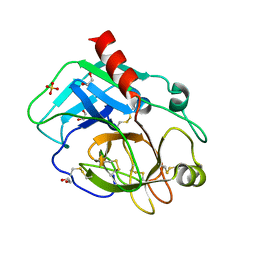 | | Structure of the KLK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Kallikrein-1, SULFATE ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of the KLK1 from Biortus.
To Be Published
|
|
8YHW
 
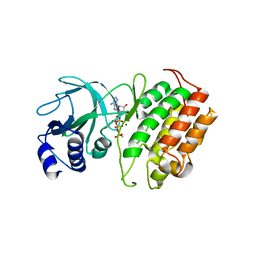 | | The Crystal Structure of NF-kB-inducing Kinase (NIK) from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, MAGNESIUM ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of NF-kB-inducing Kinase (NIK) from Biortus
To Be Published
|
|
8YHL
 
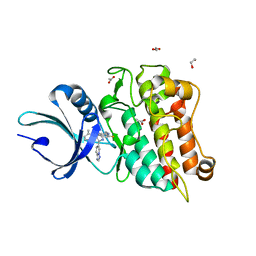 | | The Crystal Structure of Tgf-Beta Type I Receptor (Alk5) from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 2-fluoranyl-~{N}-[[5-(6-methylpyridin-2-yl)-4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-1~{H}-imidazol-2-yl]methyl]aniline, ACETATE ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of Tgf-Beta Type I Receptor (Alk5) from Biortus
To Be Published
|
|
8XVV
 
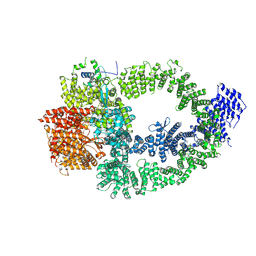 | | The TRRAP module of human NuA4/TIP60 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Isoform 2 of E1A-binding protein p400, Isoform 2 of Transformation/transcription domain-associated protein | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
8XVG
 
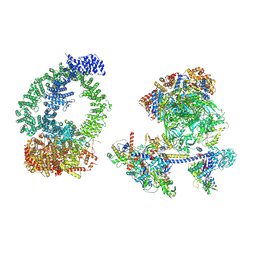 | | Structure of human NuA4/TIP60 complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, Actin-like protein 6A, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
8XVT
 
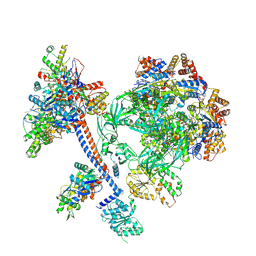 | | The core subcomplex of human NuA4/TIP60 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
6P72
 
 | | Crystal Structure of the Cedar henipavirus Attachment G Glycoprotein global domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B, Xu, Y. | | Deposit date: | 2019-06-04 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P7S
 
 | | Crystal Structure of the Cedar henipavirus Attachment G Glycoprotein globular domain in complex with the receptor ephrin-B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B, Xu, Y. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P7Y
 
 | | Crystal Structure of the Cedar henipavirus Attachment G Glycoprotein globular domain in complex with the receptor ephrin-B2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B, Xu, Y. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.844 Å) | | Cite: | Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8AZ3
 
 | | IAPP S20G growth-phase fibril polymorph 4PF-CU | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
8AZ4
 
 | | IAPP S20G plateau-phase fibril polymorph 2PF-L | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
