3GQA
 
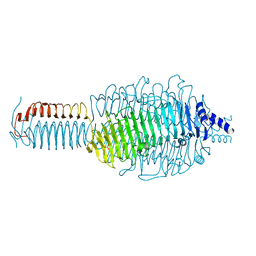 | |
5Y66
 
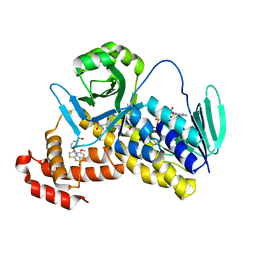 | | Crystal structure of Pseudomonas fluorescens Kynurenine 3-monooxygenase in complex with L-KYN and Ro61-8048 | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 3,4-dimethoxy-N-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]benzenesulfonamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xiang, Y, Gao, J.J, Zhu, D.Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Biochemistry and structural studies of kynurenine 3-monooxygenase reveal allosteric inhibition by Ro 61-8048
FASEB J., 32, 2018
|
|
5Y7A
 
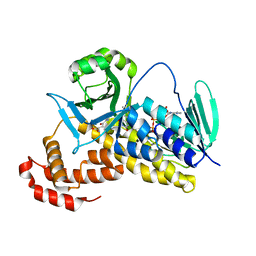 | |
5Y77
 
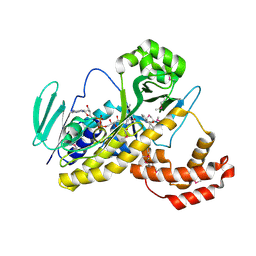 | |
8J1V
 
 | |
8J1T
 
 | |
3QC7
 
 | |
3R02
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 7-[(cis-4-aminocyclohexyl)amino]-5-bromo-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R04
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-{6-[(trans-4-aminocyclohexyl)amino]pyrazin-2-yl}-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7Y42
 
 | |
8IXJ
 
 | |
8IXK
 
 | | bottom segment of the bacteriophage M13 mini variant | | Descriptor: | Attachment protein G3P, Capsid protein G8P, Head virion protein G6P | | Authors: | Xiang, Y, Jia, Q. | | Deposit date: | 2023-04-01 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of a bacteriophage M13 mini variant.
Nat Commun, 14, 2023
|
|
8IXL
 
 | | top segment of the bacteriophage M13 mini variant | | Descriptor: | Capsid protein G8P, Tail virion protein G7P, Tail virion protein G9P | | Authors: | Xiang, Y, Jia, Q. | | Deposit date: | 2023-04-01 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of a bacteriophage M13 mini variant.
Nat Commun, 14, 2023
|
|
7WBZ
 
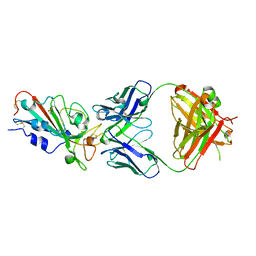 | | Crystal structure of the SARS-Cov-2 RBD in complex with Fab 2303 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2303 heavy chain, 2303 light chain, ... | | Authors: | Xiang, Y, Ma, B. | | Deposit date: | 2021-12-17 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Conformational flexibility in neutralization of SARS-CoV-2 by naturally elicited anti-SARS-CoV-2 antibodies.
Commun Biol, 5, 2022
|
|
7WC0
 
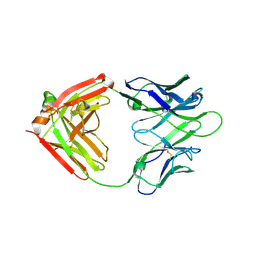 | |
7WCD
 
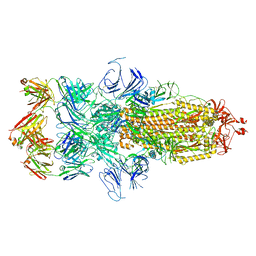 | | Cryo EM structure of SARS-CoV-2 spike in complex with TAU-2212 mAbs in conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | Authors: | Xiang, Y, Ma, B, Li, R. | | Deposit date: | 2021-12-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational flexibility in neutralization of SARS-CoV-2 by naturally elicited anti-SARS-CoV-2 antibodies.
Commun Biol, 5, 2022
|
|
1U6T
 
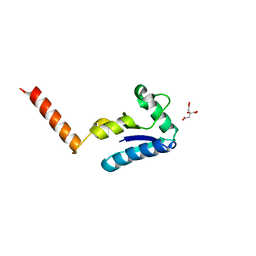 | | Crystal structure of the human SH3 binding glutamic-rich protein like | | Descriptor: | CITRIC ACID, SH3 domain-binding glutamic acid-rich-like protein | | Authors: | Yin, L, Xiang, Y, Yang, N, Zhu, D.-Y, Huang, R.-H, Wang, D.-C. | | Deposit date: | 2004-08-01 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human SH3BGRL protein: the first structure of the human SH3BGR family representing a novel class of thioredoxin fold proteins
Proteins, 61, 2005
|
|
4W60
 
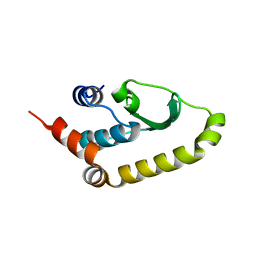 | | The structure of Vaccina virus H7 protein displays A Novel Phosphoinositide binding fold required for membrane biogenesis | | Descriptor: | Late protein H7 | | Authors: | Kolli, S, Meng, X, Wu, X, Shengjuler, D, Cameron, C.E, Xiang, Y, Deng, J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-12-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function analysis of vaccinia virus h7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication.
J.Virol., 89, 2015
|
|
5XLR
 
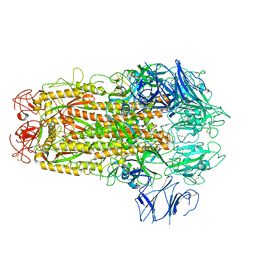 | | Structure of SARS-CoV spike glycoprotein | | Descriptor: | Spike glycoprotein | | Authors: | Gui, M, Song, W, Xiang, Y, Wang, X. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding.
Cell Res., 27, 2017
|
|
1KV0
 
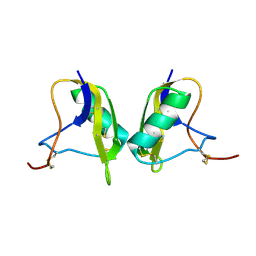 | | Cis/trans Isomerization of Non-prolyl Peptide Bond Observed in Crystal Structure of an Scorpion Toxin | | Descriptor: | Alpha-like toxin BmK-M7 | | Authors: | Guan, R.J, He, X.L, Wang, M, Xiang, Y, Wang, D.C. | | Deposit date: | 2002-01-23 | | Release date: | 2003-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural mechanism governing cis and trans isomeric states and an intramolecular switch for cis/trans isomerization of a non-proline peptide bond observed in crystal structures of scorpion toxins.
J.Mol.Biol., 341, 2004
|
|
4XFU
 
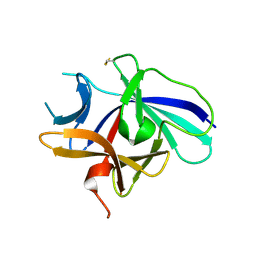 | | Structure of IL-18 SER Mutant V | | Descriptor: | Interleukin-18 | | Authors: | Krumm, B.E, Meng, X, Xiang, Y, Deng, J. | | Deposit date: | 2014-12-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallization of interleukin-18 for structure-based inhibitor design.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XFT
 
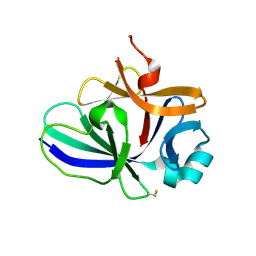 | | Structure of IL-18 SER Mutant III | | Descriptor: | DIMETHYL SULFOXIDE, Interleukin-18 | | Authors: | Krumm, B.E, Meng, X, Xiang, Y, Deng, J. | | Deposit date: | 2014-12-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization of interleukin-18 for structure-based inhibitor design.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1P9Z
 
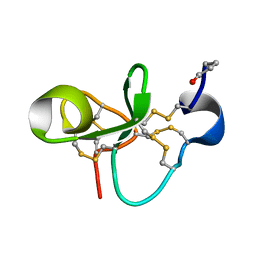 | | The Solution Structure of Antifungal Peptide Distinct With a Five-disulfide Motif from Eucommia ulmoides Oliver | | Descriptor: | Eucommia Antifungal peptide 2 | | Authors: | Huang, R.H, Xiang, Y, Tu, G.Z, Zhang, Y, Wang, D.C. | | Deposit date: | 2003-05-13 | | Release date: | 2004-05-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Eucommia Antifungal Peptide: A Novel Structural Model Distinct with a Five-Disulfide Motif.
Biochemistry, 43, 2004
|
|
5WRG
 
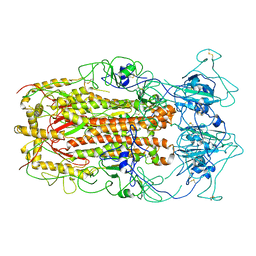 | | SARS-CoV spike glycoprotein | | Descriptor: | Spike glycoprotein | | Authors: | Gui, M, Song, W, Xiang, Y, Wang, X. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding.
Cell Res., 27, 2017
|
|
6QX7
 
 | |
