6M0X
 
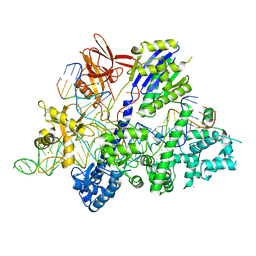 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with AGGA PAM | | Descriptor: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | Authors: | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | Deposit date: | 2020-02-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
7X1Z
 
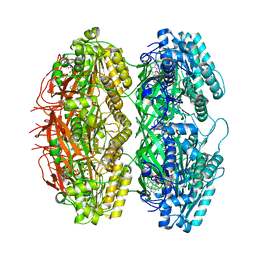 | | Structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
7X1Y
 
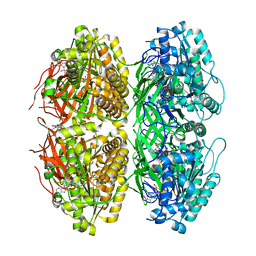 | | Structure of the phosphorylation-site double mutant S431A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
8ZC9
 
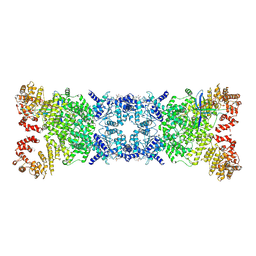 | | The Cryo-EM structure of DSR2-Tail tube-NAD+ complex | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein, tail tube protein | | Authors: | Wang, R, Xu, Q, Wu, Z, Li, J, Yang, R, Shi, Z, Li, F. | | Deposit date: | 2024-04-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
6LTH
 
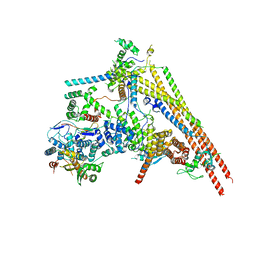 | | Structure of human BAF Base module | | Descriptor: | AT-rich interactive domain-containing protein 1A, SWI/SNF complex subunit SMARCC2, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1, ... | | Authors: | He, S, Wu, Z, Tian, Y, Yu, Z, Yu, J, Wang, X, Li, J, Liu, B, Xu, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of nucleosome-bound human BAF complex.
Science, 367, 2020
|
|
6LTJ
 
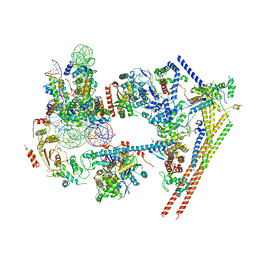 | | Structure of nucleosome-bound human BAF complex | | Descriptor: | AT-rich interactive domain-containing protein 1A, Actin, cytoplasmic 1, ... | | Authors: | He, S, Wu, Z, Tian, Y, Yu, Z, Yu, J, Wang, X, Li, J, Liu, B, Xu, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of nucleosome-bound human BAF complex.
Science, 367, 2020
|
|
7BWA
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 2 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW7
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin. | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-13 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW8
 
 | | Cryo-EM Structure for the Insulin Binding Region in the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
8Y3Y
 
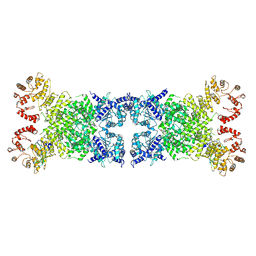 | | The Cryo-EM structure of anti-phage defense associated DSR2 tetramer bound with two DSAD1 inhibitors (opposite side) | | Descriptor: | DSR anti-defence 1, SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y34
 
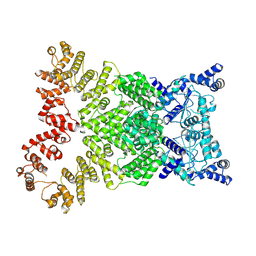 | | Cryo-EM structure of anti-phage defense associated DSR2 (H171A) (map2) | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-28 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
7X63
 
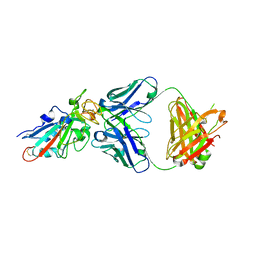 | | SARS-CoV-2-Beta-RBD and BD-236-GWP/P-VK antibody complex | | Descriptor: | BD-236 Fab heavy chain, BD-236 Fab light chain, Spike protein S1 | | Authors: | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | Deposit date: | 2022-03-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | SARS-CoV-2-Beta-RBD and BD-236-GWP/P-VK antibody complex
To Be Published
|
|
7XIL
 
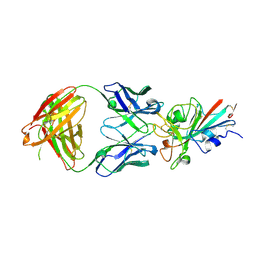 | | SARS-CoV-2-Beta-RBD and B38-GWP/P-VK antibody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B38 Fab heavy chain, B38 Fab light chain, ... | | Authors: | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | SARS-CoV-2-Beta-RBD and B38-GWP/P-VK antibody complex
To Be Published
|
|
7XJZ
 
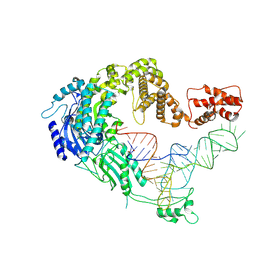 | | Cryo-EM strucrture of Oryza sativa plastid glycyl-tRNA synthetase in complex with tRNA (tRNA binding state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XJY
 
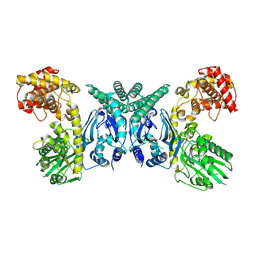 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase (apo form) | | Descriptor: | Glycine--tRNA ligase | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XK1
 
 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase in complex with two tRNAs (both in tRNA binding states) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XK0
 
 | | Cryo-EM strucrture of Oryza sativa plastid glycyl-tRNA synthetase in complex with tRNA (tRNA locked state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7X66
 
 | | SARS-CoV-2-Omicron-RBD and BD-236-GWP/P-VK antibody complex | | Descriptor: | BD-236 Fab heavy chain, BD-236 Fab light chain, Spike protein S1 | | Authors: | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | Deposit date: | 2022-03-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2-Omicron-RBD and BD-236-GWP/P-VK antibody complex
To Be Published
|
|
7XIK
 
 | | SARS-CoV-2-Omicron-RBD and B38-GWP/P-VK antibody complex | | Descriptor: | B38 Fab heavy chain, B38 Fab light chain, Spike protein S1 | | Authors: | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | SARS-CoV-2-Omicron-RBD and B38-GWP/P-VK antibody complex
To Be Published
|
|
7XUK
 
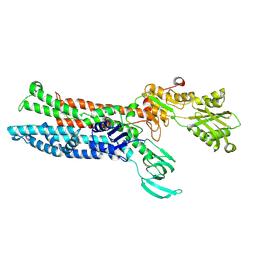 | |
7XUM
 
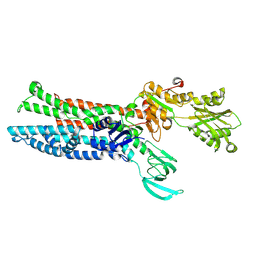 | | Structure of ATP7B C983S/C985S/D1027A mutant with Cu+ in presence of ATOX1 | | Descriptor: | COPPER (II) ION, Copper-transporting ATPase 2 | | Authors: | Yang, G, Xu, L, Chang, S, Guo, J, Wu, Z. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
7XUN
 
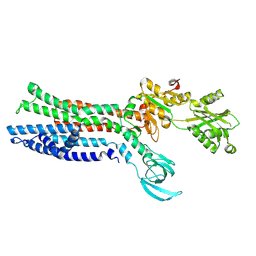 | |
7XUO
 
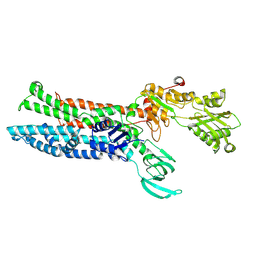 | | Structure of ATP7B C983S/C985S/D1027A mutant with cisplatin in presence of ATOX1 | | Descriptor: | Copper-transporting ATPase 2, PLATINUM (II) ION | | Authors: | Yang, G, Xu, L, Chang, S, Guo, J, Wu, Z. | | Deposit date: | 2022-05-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
8K3B
 
 | |
1YOP
 
 | | The solution structure of Kti11p | | Descriptor: | Kti11p, ZINC ION | | Authors: | Sun, J, Zhang, J, Wu, F, Xu, C, Li, S, Zhao, W, Wu, Z, Wu, J, Zhou, C.-Z, Shi, Y. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module.
Biochemistry, 44, 2005
|
|
