4LLY
 
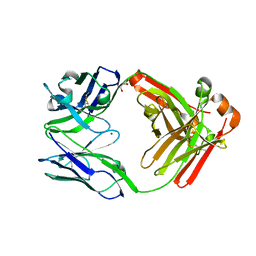 | | Crystal structure of Pertuzumab Clambda Fab with variable and constant domain redesigns (VRD2 and CRD2) at 1.6A | | Descriptor: | GLYCEROL, MAGNESIUM ION, light chain Clambda, ... | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLQ
 
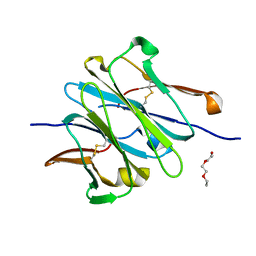 | | Structure of redesigned IgG1 first constant and lambda domains (CH1:Clambda constant redesign 2 beta, CRD2b) at 1.42A | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, mutated CH1, mutated light chain Clambda | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLU
 
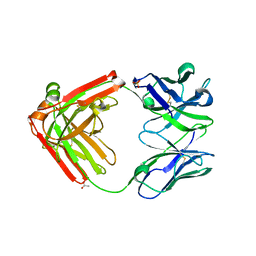 | | Structure of Pertuzumab Fab with light chain Clambda at 2.16A | | Descriptor: | ACETATE ION, Light chain CLAMBDA, PERTUZUMAB FAB Heavy chain, ... | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLM
 
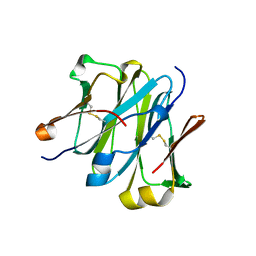 | | Structure of redesigned IgG1 first constant and lambda domains (CH1:Clambda constant redesign 1, CRD1) at 1.75A | | Descriptor: | Ig gamma-1 chain C region, Ig lambda-2 chain C region | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4JZA
 
 | |
7CFM
 
 | | Cryo-EM structure of the P395-bound GPBAR-Gs complex | | Descriptor: | 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5H-pyrido[4,3-d]pyrimidine-4-carboxamide, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
7XIJ
 
 | | Crystal structure of CBP bromodomain liganded with Y08175 | | Descriptor: | 3-[(1-ethanoyl-5-methoxy-indol-3-yl)carbonylamino]-4-fluoranyl-5-(1-methylpyrazol-4-yl)benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08175
To Be Published
|
|
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | Descriptor: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
4P42
 
 | | Extended-Synaptotagmin 2, SMP - C2A - C2B Domains | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, Extended synaptotagmin-2 | | Authors: | Schauder, C.M, Wu, X, Reinisch, K.M. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer.
Nature, 510, 2014
|
|
8TNL
 
 | |
8TOA
 
 | | CryoEM structure of H7 hemagglutinin from A/Shanghai2/2013 H7N9 in complex with a human neutralizing antibody H7.HK2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H7.HK2 Neutralizing Antibody Heavy Chain, H7.HK2 Neutralizing Antibody Light Chain, ... | | Authors: | Morano, N.C, Becker, J.E, Wu, X, Shapiro, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | CryoEM structure of H7 hemagglutinin from A/Shanghai2/2013 H7N9 in complex with a human neutralizing antibody H7.HK1
To Be Published
|
|
7EPT
 
 | | Structural basis for the tethered peptide activation of adhesion GPCRs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ping, Y.-Q, Xiao, P, Yang, F, Zhao, R.-J, Guo, S.-C, Yan, X, Wu, X, Liebscher, I, Xu, H.E, Sun, J.-P. | | Deposit date: | 2021-04-27 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
6JV0
 
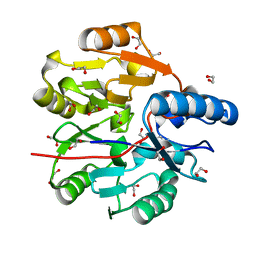 | | Crystal Structure of N-terminal domain of ArgZ, bound to Product, an arginine dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | 1,2-ETHANEDIOL, L-ornithine, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
6JUY
 
 | | Crystal Structure of ArgZ, apo structure, an Arginine Dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
6JV1
 
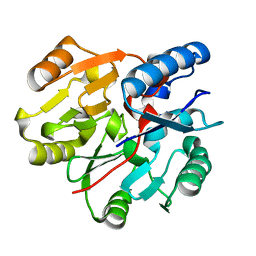 | | Crystal Structure of N-terminal domain of ArgZ, C264S mutant, bound to Substrate, an arginine dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | ARGININE, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
6JUZ
 
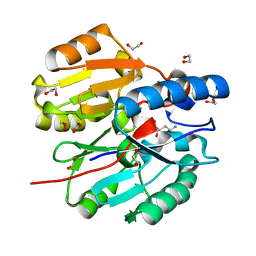 | | Crystal Structure of N-terminal domain of ArgZ(N71S) covalently bond to a reaction intermediate | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhuang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
6LCP
 
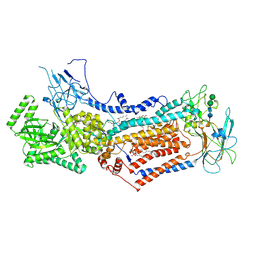 | | Cryo-EM structure of Dnf1 from Chaetomium thermophilum in the E2P state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, Y, Xu, J, Wu, X, Li, L. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structures of a P4-ATPase lipid flippase in lipid bilayers.
Protein Cell, 11, 2020
|
|
7WHW
 
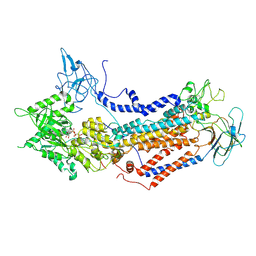 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in detergent with AMPPCP (E1-ATP state) | | Descriptor: | Alkylphosphocholine resistance protein LEM3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7WHV
 
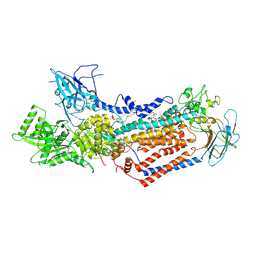 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in detergent with beryllium fluoride (E2P state) | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7F7F
 
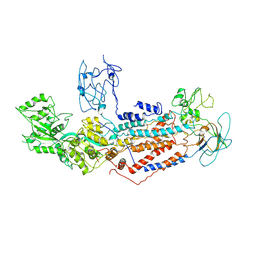 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in yeast lipids with beryllium fluoride (resting state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-06-29 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DRX
 
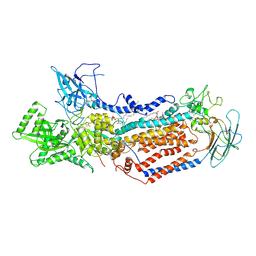 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with beryllium fluoride (E2P state) | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-30 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DSH
 
 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with AMPPCP (E1-ATP state) | | Descriptor: | Alkylphosphocholine resistance protein LEM3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DSI
 
 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in yeast lipids with AMPPCP ( resting state ) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
