4D2P
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 7-({4-[(3-hydroxy-5-methoxyphenyl)amino]benzoyl}amino)-1,2,3,4-tetrahydroisoquinolinium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
3MRU
 
 | | Crystal Structure of Aminoacylhistidine Dipeptidase from Vibrio alginolyticus | | Descriptor: | Aminoacyl-histidine dipeptidase, ZINC ION | | Authors: | Chang, C.-Y, Hsieh, Y.-C, Wu, T.-K, Chen, C.-J. | | Deposit date: | 2010-04-29 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and mutational analysis of aminoacylhistidine dipeptidase from vibrio alginolyticus reveal a new architecture of M20 metallopeptidases
J.Biol.Chem., 285, 2010
|
|
6PYF
 
 | | Sex Hormone-binding globulin mutant E176K in complex with Estradiol | | Descriptor: | CALCIUM ION, ESTRADIOL, Sex hormone-binding globulin | | Authors: | Round, P.W, Wu, T.S, Das, S, Van Petegem, F. | | Deposit date: | 2019-07-29 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular interactions between sex hormone-binding globulin and nonsteroidal ligands that enhance androgen activity.
J.Biol.Chem., 295, 2020
|
|
3AMR
 
 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CALCIUM ION, D-MYO-INOSITOL-HEXASULPHATE | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
3AMS
 
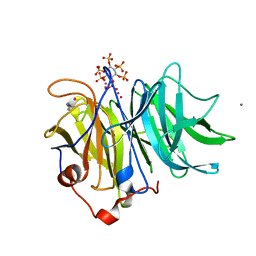 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Cd2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CADMIUM ION, CALCIUM ION, ... | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
8X91
 
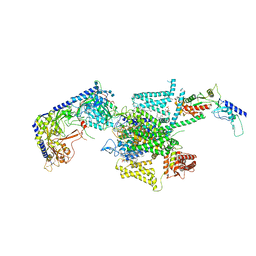 | | P/Q type calcium channel in complex with omega-conotoxin MVIIC | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Cong, Y, Wu, T, Wang, T. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for different omega-agatoxin IVA sensitivities of the P-type and Q-type Ca v 2.1 channels.
Cell Res., 34, 2024
|
|
8X90
 
 | | P/Q type calcium channel | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-[PHOSPHO-L-SERINE], 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Cong, Y, Wu, T, Wang, T. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for different omega-agatoxin IVA sensitivities of the P-type and Q-type Ca v 2.1 channels.
Cell Res., 34, 2024
|
|
8X93
 
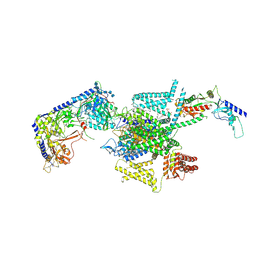 | | P/Q type calcium channel in complex with omega-Agatoxin IVA | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Cong, Y, Wu, T, Wang, T. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis for different omega-agatoxin IVA sensitivities of the P-type and Q-type Ca v 2.1 channels.
Cell Res., 34, 2024
|
|
3WDY
 
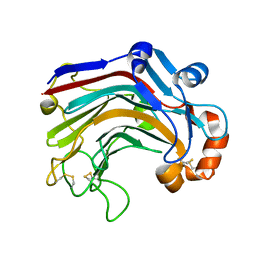 | | The complex structure of E113A with cellotetraose | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION, beta-D-glucopyranose, ... | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WDT
 
 | | The apo-form structure of PtLic16A from Paecilomyces thermophila | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WDU
 
 | | The complex structure of PtLic16A with cellobiose | | Descriptor: | Beta-1,3-1,4-glucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WH9
 
 | | The ligand-free structure of ManBK from Aspergillus niger BK01 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, J.W, Chen, C.C, Huang, C.H, Huang, T.Y, Wu, T.H, Cheng, Y.S, Ko, T.P, Lin, C.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Analysis and Rational Design to Improve Specific Activity of beta-Mannanase from Aspergillus Niger BK01
To be Published
|
|
3WDW
 
 | | The apo-form structure of E113A from Paecilomyces thermophila | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WDV
 
 | | The complex structure of PtLic16A with cellotetraose | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION, beta-D-glucopyranose, ... | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WDX
 
 | | The complex structure of E113A with glucotriose | | Descriptor: | Beta-1,3-1,4-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
1C72
 
 | | TYR115, GLN165 AND TRP209 CONTRIBUTE TO THE 1,2-EPOXY-3-(P-NITROPHENOXY)PROPANE CONJUGATING ACTIVITIES OF GLUTATHIONE S-TRANSFERASE CGSTM1-1 | | Descriptor: | 1-HYDROXY-2-S-GLUTATHIONYL-3-PARA-NITROPHENOXY-PROPANE, PROTEIN (GLUTATHIONE S-TRANSFERASE) | | Authors: | Chern, M.K, Wu, T.C, Hsieh, C.H, Chou, C.C, Liu, L.F, Kuan, I.C, Yeh, Y.H, Hsiao, C.D, Tam, M.F. | | Deposit date: | 2000-02-02 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tyr115, gln165 and trp209 contribute to the 1, 2-epoxy-3-(p-nitrophenoxy)propane-conjugating activity of glutathione S-transferase cGSTM1-1.
J.Mol.Biol., 300, 2000
|
|
7XIJ
 
 | | Crystal structure of CBP bromodomain liganded with Y08175 | | Descriptor: | 3-[(1-ethanoyl-5-methoxy-indol-3-yl)carbonylamino]-4-fluoranyl-5-(1-methylpyrazol-4-yl)benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08175
To Be Published
|
|
7XNE
 
 | | Crystal structure of CBP bromodomain liganded with Y08284 | | Descriptor: | CREB-binding protein, GLYCEROL, N-[3-(1-cyclopropylpyrazol-4-yl)-2-fluoranyl-5-[(1S)-1-oxidanylethyl]phenyl]-3-ethanoyl-7-methoxy-indolizine-1-carboxamide | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, C, Hu, Q, Luo, G, Hu, J, Zhuang, X, Zou, L, Shen, H, Wu, X, Zhang, Y, Kong, X, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
2PK4
 
 | |
8GXD
 
 | | L-LEUCINE DEHYDROGENASE FROM EXIGUOBACTERIUM SIBIRICUM | | Descriptor: | CALCIUM ION, GLYCEROL, Glu/Leu/Phe/Val dehydrogenase | | Authors: | Mu, X, Nie, Y, Wu, T, Wang, Y, Zhang, N, Yin, D, Xu, Y. | | Deposit date: | 2022-09-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Reshaping Substrate-Binding Pocket of Leucine Dehydrogenase for Bidirectionally Accessing Structurally Diverse Substrates
Acs Catalysis, 13, 2023
|
|
8I78
 
 | | Meso-Diaminopimelate dehydrogenase | | Descriptor: | Meso-diaminopimelate D-dehydrogenase | | Authors: | Wei, S, Wu, T.F. | | Deposit date: | 2023-01-31 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Analysis of the catalytic mechanism of meso-DAPDH and extension of D-aromatic amino acid substrate scope
To Be Published
|
|
1P2F
 
 | |
7EVJ
 
 | | Crystal structure of CBP bromodomain liganded with 9c | | Descriptor: | 3-acetyl-1-((3-(1-cyclopropyl-1H-pyrazol-4-yl)-2-fluoro-5-(hydroxymethyl)phenyl)carbamoyl)indolizin-7-yl dimethylcarbamate, CREB-binding protein, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
1ZH2
 
 | |
