3WGO
 
 | | Crystal structure of Q154L/T173I/R199M/P248S/H249/N276S mutant of meso-dapdh from Clostridium tetani E88 | | 分子名称: | Meso-diaminopimelate dehydrogenase | | 著者 | Liu, W.D, Li, Z, Huang, C.H, Guo, R.T, Wu, Q.Q, Zhu, D.M. | | 登録日 | 2013-08-06 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of Q154L/T173I/R199M/P248S/H249 mutant of meso-dapdh from Clostridium tetani E88
To be Published
|
|
2BYC
 
 | | BlrB - a BLUF protein, dark state structure | | 分子名称: | BLUE-LIGHT RECEPTOR OF THE BLUF-FAMILY, FLAVIN MONONUCLEOTIDE | | 著者 | Jung, A, Domratcheva, T, Tarutina, M, Wu, Q, Ko, W.H, Shoeman, R.L, Gomelsky, M, Gardner, K.H, Schlichting, I. | | 登録日 | 2005-07-29 | | 公開日 | 2005-08-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of a Bacterial Bluf Photoreceptor: Insights Into Blue Light-Mediated Signal Transduction.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2AN3
 
 | | Structure of PNMT with S-adenosyl-L-homocysteine and the semi-rigid analogue acceptor substrate cis-(1R,2S)-2-amino-1-tetralol. | | 分子名称: | CIS-(1R,2S)-2-AMINO-1,2,3,4-TETRAHYDRONAPHTHALEN-1-OL, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | 登録日 | 2005-08-11 | | 公開日 | 2006-03-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
2AN5
 
 | | Structure of human PNMT complexed with S-adenosyl-homocysteine and an inhibitor, trans-(1S,2S)-2-amino-1-tetralol | | 分子名称: | PHOSPHATE ION, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | 登録日 | 2005-08-11 | | 公開日 | 2006-03-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
2AN4
 
 | | Structure of PNMT complexed with S-adenosyl-L-homocysteine and the acceptor substrate octopamine | | 分子名称: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, PHOSPHATE ION, Phenylethanolamine N-methyltransferase, ... | | 著者 | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | 登録日 | 2005-08-11 | | 公開日 | 2006-03-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
2F9S
 
 | | 2nd Crystal Structure Of A Soluble Domain Of ResA In The Oxidised Form | | 分子名称: | Thiol-disulfide oxidoreductase resA | | 著者 | Colbert, C.L, Wu, Q, Erbel, P.J.A, Gardner, K.H, Deisenhofer, J. | | 登録日 | 2005-12-06 | | 公開日 | 2006-04-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.401 Å) | | 主引用文献 | Mechanism of substrate specificity in Bacillus subtilis ResA, a thioredoxin-like protein involved in cytochrome c maturation
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
7MK3
 
 | | Crystal structure of NPR1 | | 分子名称: | CHLORIDE ION, GLYCEROL, Regulatory protein NPR1, ... | | 著者 | Cheng, J, Wu, Q, Zhou, P. | | 登録日 | 2021-04-21 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
4JGV
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with THPN | | 分子名称: | 1-(3,4,5-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | 著者 | Zhang, Q, Li, F, Li, A, Tian, X, Wan, W, Wan, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | 登録日 | 2013-03-04 | | 公開日 | 2013-12-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4RZF
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, S441W mutant | | 分子名称: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | 著者 | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | 登録日 | 2014-12-21 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
8HQP
 
 | | Crystal structure of AbHheG mutant from Acidimicrobiia bacterium | | 分子名称: | AbHheG_m | | 著者 | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | 登録日 | 2022-12-13 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | 分子名称: | GLYCEROL, alpha/beta hydrolase | | 著者 | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | 登録日 | 2022-10-24 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
1K88
 
 | | Crystal structure of procaspase-7 | | 分子名称: | procaspase-7 | | 著者 | Chai, J, Wu, Q, Shiozaki, E, Srinivasa, S.M, Alnemri, E.S, Shi, Y. | | 登録日 | 2001-10-23 | | 公開日 | 2001-11-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding
Cell(Cambridge,Mass.), 107, 2001
|
|
1K86
 
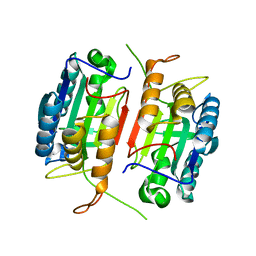 | | Crystal structure of caspase-7 | | 分子名称: | caspase-7 | | 著者 | Chai, J, Wu, Q, Shiozaki, E, Srinivasa, S.M, Alnemri, E.S, Shi, Y. | | 登録日 | 2001-10-23 | | 公開日 | 2001-11-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding
Cell(Cambridge,Mass.), 107, 2001
|
|
8JQK
 
 | | Crystal structure of a carbonyl reductase SSCR mutant from Sporobolomyces Salmonicolor | | 分子名称: | Aldehyde reductase 2 | | 著者 | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | 登録日 | 2023-06-14 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
8JQJ
 
 | | Crystal structure of carbonyl reductase SSCR mutant 1 from Sporobolomyces Salmonicolor | | 分子名称: | Aldehyde reductase 2 | | 著者 | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | 登録日 | 2023-06-14 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
3VNN
 
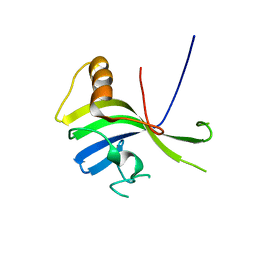 | | Crystal Structure of a sub-domain of the nucleotidyltransferase (adenylation) domain of human DNA ligase IV | | 分子名称: | DNA ligase 4 | | 著者 | Ochi, T, Wu, Q, Chirgadze, D.Y, Grossmann, J.G, Bolanos-Garcia, V.M, Blundell, T.L. | | 登録日 | 2012-01-17 | | 公開日 | 2012-06-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.903 Å) | | 主引用文献 | Structural insights into the role of domain flexibility in human DNA ligase IV
Structure, 20, 2012
|
|
6J1Q
 
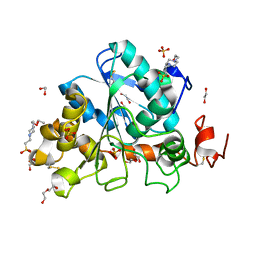 | | Crystal structure of Candida Antarctica Lipase B mutant - RS | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Cen, Y.X, Zhou, J.H, Wu, Q. | | 登録日 | 2018-12-29 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
6J1T
 
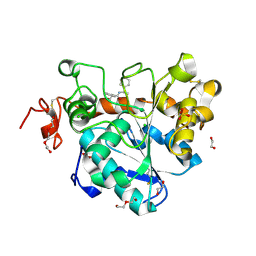 | | Crystal structure of Candida Antarctica Lipase B mutant SR with product analogue | | 分子名称: | (2S)-2-phenyl-N-[(1R)-1-phenylethyl]propanamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Cen, Y.X, Zhou, J.H, Wu, Q. | | 登録日 | 2018-12-29 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.783 Å) | | 主引用文献 | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
4RE8
 
 | | Crystal Structure of TR3 LBD in complex with Molecule 5 | | 分子名称: | 1-(3,4,5-trihydroxyphenyl)dodecan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | 著者 | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Weijia, W, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | 登録日 | 2014-09-22 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
6J1P
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - SR | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Cen, Y.X, Zhou, J.H, Wu, Q. | | 登録日 | 2018-12-29 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.759 Å) | | 主引用文献 | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
4REE
 
 | | Crystal Structure of TR3 LBD in complex with Molecule 6 | | 分子名称: | 1-(2,3,4-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | 著者 | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | 登録日 | 2014-09-22 | | 公開日 | 2015-09-09 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4REF
 
 | | Crystal Structure of TR3 LBD_L449W in complex with Molecule 2 | | 分子名称: | 1-(3,4,5-trihydroxyphenyl)hexan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | 著者 | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | 登録日 | 2014-09-22 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
3P4K
 
 | | The third conformation of p38a MAP kinase observed in phosphorylated p38a and in solution | | 分子名称: | MAP kinase 14, Mitogen-activated protein kinase 14 | | 著者 | Akella, R, Min, X, Wu, Q, Gardner, K.H, Goldsmith, E.J. | | 登録日 | 2010-10-06 | | 公開日 | 2011-01-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.304 Å) | | 主引用文献 | The third conformation of p38a MAP kinase observed in phosphorylated p38a and in solution
Structure, 18, 2010
|
|
2MNC
 
 | |
5GGM
 
 | | The NMR structure of calmodulin in CTAB reverse micelles | | 分子名称: | CALCIUM ION, Calmodulin, TERBIUM(III) ION | | 著者 | Xu, G, Cheng, K, Wu, Q, Liu, M, Li, C. | | 登録日 | 2016-06-16 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR structure of calmodulin in CTAB reverse micelles
To Be Published
|
|
