6AOV
 
 | |
7JMW
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain in complex with cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
7JMX
 
 | | Crystal structure of a SARS-CoV-2 cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | ACETATE ION, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
6XC4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 heavy chain, CC12.3 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC3
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.1 and CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.3 and CR3022 | | Descriptor: | CC12.3 heavy chain, CC12.3 light chain, CR3022 heavy chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC2
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.112 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
7LKA
 
 | | Crystal structure of SARS-CoV-2 RBD-targeting antibody COV107-23 | | Descriptor: | ACETATE ION, COV107-23 heavy chain, COV107-23 light chain | | Authors: | Yuan, M, Zhu, X, Wilson, I.A, Wu, N.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sequence signatures of two public antibody clonotypes that bind SARS-CoV-2 receptor binding domain.
Nat Commun, 12, 2021
|
|
7LK9
 
 | | Crystal structure of SARS-CoV-2 RBD-targeting antibody COV107-23 HC + COVD21-C8 LC | | Descriptor: | COV107-23 heavy chain, COVD21-C8 light chain | | Authors: | Yuan, M, Zhu, X, Wilson, I.A, Wu, N.C. | | Deposit date: | 2021-02-01 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Sequence signatures of two public antibody clonotypes that bind SARS-CoV-2 receptor binding domain.
Nat Commun, 12, 2021
|
|
6W41
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab heavy chain, CR3022 Fab light chain, ... | | Authors: | Yuan, M, Wu, N.C, Zhu, X.Y, Wilson, I.A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.084 Å) | | Cite: | A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV.
Science, 368, 2020
|
|
6XKP
 
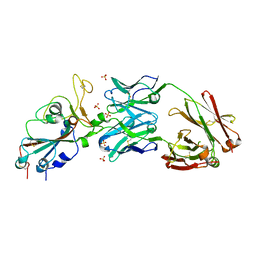 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-270 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-270 Heavy Chain, CV07-270 Light Chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
6XKQ
 
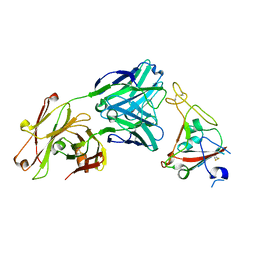 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-250 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-250 Heavy Chain, CV07-250 Light Chain, ... | | Authors: | Yuan, M, Liu, H, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
7KN5
 
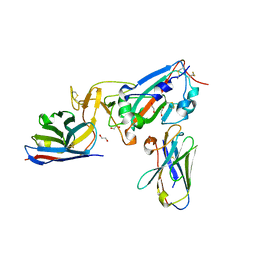 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobodies VHH E and U | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7KN7
 
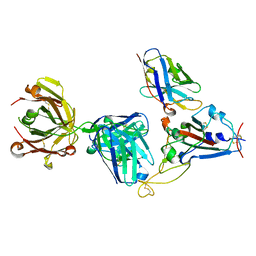 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobody VHH W and antibody Fab CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, CC12.3 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7KN6
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobody VHH V and antibody Fab CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, CC12.3 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
8EZ8
 
 | | Structure of 3C08 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 3C08, Light chain of influenza virus neuraminidase antibody 3C08, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase.
Immunity, 56, 2023
|
|
8EZ7
 
 | | Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 1F04, Light chain of influenza virus neuraminidase antibody 1F04, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase.
Immunity, 56, 2023
|
|
8EZ3
 
 | | Structure of 3A10 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 3A10, Light chain of influenza virus neuraminidase antibody 3A10, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase.
Immunity, 56, 2023
|
|
