7WTE
 
 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the intermediate state 2 | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-TRIPHOSPHATE, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTA
 
 | | Cryo-EM structure of human pyruvate carboxylase in apo state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Pyruvate carboxylase, mitochondrial | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTC
 
 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the ground state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTB
 
 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
4JCL
 
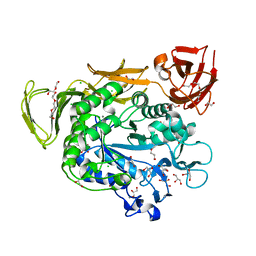 | | Crystal structure of Alpha-CGT from Paenibacillus macerans at 1.7 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J, Wu, J, Li, J, Chen, J. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Alpha-Cgt from Paenibacillus Macerans at 1.7 Angstrom Resolution
To be Published
|
|
8CMN
 
 | |
8U4E
 
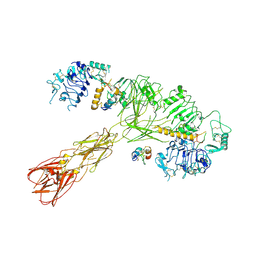 | | Cryo-EM structure of long form insulin receptor (IR-B) with three IGF2 bound, asymmetric conformation. | | Descriptor: | Insulin receptor, Insulin-like growth factor II | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8U4B
 
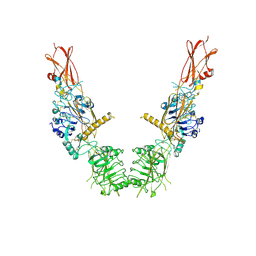 | | Cryo-EM structure of long form insulin receptor (IR-B) in the apo state | | Descriptor: | Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8U4C
 
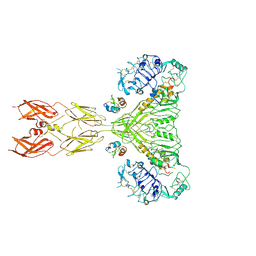 | | Cryo-EM structure of long form insulin receptor (IR-B) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin receptor, Insulin-like growth factor II | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
2A5M
 
 | |
2QUR
 
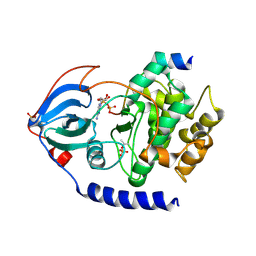 | | Crystal Structure of F327A/K285P Mutant of cAMP-dependent Protein Kinase | | Descriptor: | 20-mer fragment from cAMP-dependent protein kinase inhibitor alpha, ADENOSINE-5'-DIPHOSPHATE, cAMP-dependent protein kinase, ... | | Authors: | Taylor, S.S, Yang, J, Wu, J. | | Deposit date: | 2007-08-06 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contribution of non-catalytic core residues to activity and regulation in protein kinase A.
J.Biol.Chem., 284, 2009
|
|
2LNW
 
 | | Identification and structural basis for a novel interaction between Vav2 and Arap3 | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3, Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
2LNX
 
 | | Solution structure of Vav2 SH2 domain | | Descriptor: | Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
5JR7
 
 | | Crystal structure of an ACRDYS heterodimer [RIa(92-365):C] of PKA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Bruystens, J.G.H, Wu, J, Taylor, S.S. | | Deposit date: | 2016-05-05 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structure of a PKA RI alpha Recurrent Acrodysostosis Mutant Explains Defective cAMP-Dependent Activation.
J. Mol. Biol., 428, 2016
|
|
6W9E
 
 | | Crystal Structure of Human CDK9/cyclinT1 in complex with MC180295 | | Descriptor: | (4-amino-2-{[(1R,2R,4S)-bicyclo[2.2.1]heptan-2-yl]amino}-1,3-thiazol-5-yl)(2-nitrophenyl)methanone, (4-amino-2-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]amino}-1,3-thiazol-5-yl)(2-nitrophenyl)methanone, Cyclin-T1, ... | | Authors: | Zhang, P, Wu, J. | | Deposit date: | 2020-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Comparative Modeling of CDK9 Inhibitors to Explore Selectivity and Structure-Activity Relationships
To Be Published
|
|
5O35
 
 | | Structure of complement proteins complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, Complement factor H,Complement factor H | | Authors: | Xue, X, Wu, J, Forneris, F, Gros, P. | | Deposit date: | 2017-05-23 | | Release date: | 2017-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4JCM
 
 | | Crystal structure of Gamma-CGTASE from Alkalophilic bacillus clarkii at 1.65 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, L, Yang, D, Zhou, J, Wu, J, Chen, J. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Gamma-Cgtase from Alkalophilic Bacillus Clarkii at 1.65 Angstrom Resolution.
To be Published
|
|
5O32
 
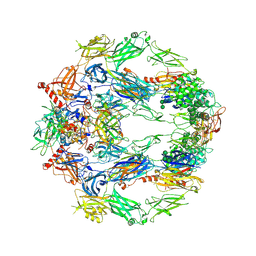 | | The structure of complement complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xue, X, Wu, J, Forneris, F, Gros, P. | | Deposit date: | 2017-05-23 | | Release date: | 2017-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.206209 Å) | | Cite: | Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5JI8
 
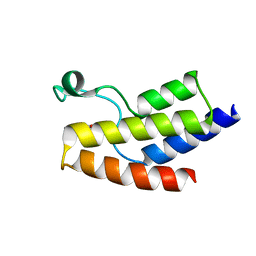 | | Crystal structure of the BRD9 bromodomain and hit 1 | | Descriptor: | 2-amino-1,3-benzothiazole-6-carboxamide, Bromodomain-containing protein 9 | | Authors: | Wang, N, Li, F, Bao, H, Li, J, Wu, J, Ruan, K. | | Deposit date: | 2016-04-22 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | NMR Fragment Screening Hit Induces Plasticity of BRD7/9 Bromodomains
Chembiochem, 17, 2016
|
|
5KE4
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 6 nicotinic acetylcholine receptor in complex with 2-((5-(3,7-Diazabicyclo[3.3.1]nonan-3-yl)pyridin-3-yl)oxy)- N,N-dimethylethanamine (BPC) | | Descriptor: | 2-((5-(3,7-Diazabicyclo[3.3.1]nonan-3-yl)pyridin-3-yl)oxy)-N,N-dimethylethanamine, Soluble acetylcholine receptor | | Authors: | Bobango, J, Wu, J, Talley, I.T, Ralston, R, Sankaran, B, Talley, T.T. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 6 nicotinic acetylcholine receptor in complex with 2-((5-(3,7-Diazabicyclo[3.3.1]nonan-3-yl)pyridin-3-yl)oxy)-
N,N-dimethylethanamine (BPC)
To Be Published
|
|
5H65
 
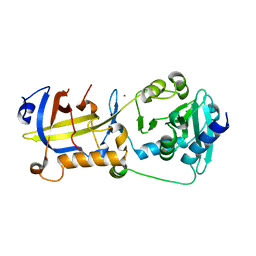 | | Crystal structure of human POT1 and TPP1 | | Descriptor: | Adrenocortical dysplasia protein homolog, Protection of telomeres protein 1, ZINC ION | | Authors: | Chen, C, Wu, J, Lei, M. | | Deposit date: | 2016-11-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer.
Nat Commun, 8, 2017
|
|
5KZU
 
 | | Crystal structure of an acetylcholine binding protein from Aplysia californica (Ac-AChBP) in complex with click chemistry compound 9-[[1-[8-methyl-8-(2-phenylethyl)-8-azoniabicyclo[3.2.1]octan-3-yl]triazol-4-yl]methyl]carbazole | | Descriptor: | 9-[[1-[8-methyl-8-(2-phenylethyl)-8-azoniabicyclo[3.2.1]octan-3-yl]triazol-4-yl]methyl]carbazole, SULFATE ION, Soluble acetylcholine receptor, ... | | Authors: | Bobango, J, Wu, J, Talley, I.T, Sankaran, B, Talley, T.T. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) in complex with click chemistry compound 9-[[1-[8-methyl-8-(2-phenylethyl)-8-azoniabicyclo[3.2.1]octan-3-yl]triazol-4-yl]methyl]carbazole
To Be Published
|
|
7LZ4
 
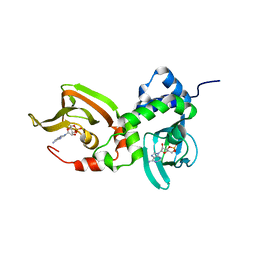 | | Crystal structure of A211D mutant of Protein Kinase A RIa subunit, a Carney Complex mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit, N-terminally processed | | Authors: | Del Rio, J, Wu, J, Taylor, S.S. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.155 Å) | | Cite: | Noncanonical protein kinase A activation by oligomerization of regulatory subunits as revealed by inherited Carney complex mutations.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2ZN7
 
 | | CRYSTAL STRUCTURES OF PTP1B-Inhibitor Complexes | | Descriptor: | 4-bromo-3-(carboxymethoxy)-5-{3-[cyclohexyl(phenylcarbonyl)amino]phenyl}thiophene-2-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Wu, J. | | Deposit date: | 2008-04-22 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase-1 B inhibitors: capturing interactions with arginine 24
Chemmedchem, 3, 2008
|
|
4O26
 
 | |
