3SE7
 
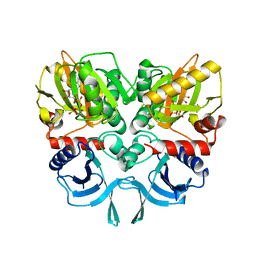 | | ancient VanA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, VanA | | Authors: | Wright, G.D, Morar, M. | | Deposit date: | 2011-06-10 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Antibiotic resistance is ancient.
Nature, 477, 2011
|
|
4WQK
 
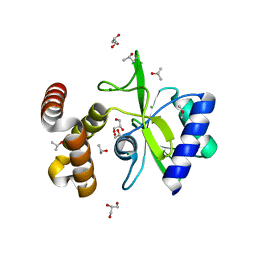 | | Crystal structure of aminoglycoside nucleotidylyltransferase ANT(2")-Ia, apo form | | Descriptor: | 2''-aminoglycoside nucleotidyltransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cox, G, Stogios, P.J, Savchenko, A, Wright, G.D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Structural and Molecular Basis for Resistance to Aminoglycoside Antibiotics by the Adenylyltransferase ANT(2)-Ia.
Mbio, 6, 2015
|
|
4WQL
 
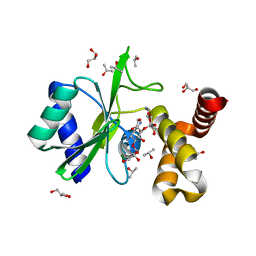 | | Crystal structure of aminoglycoside nucleotidylyltransferase ANT(2")-Ia, kanamycin-bound | | Descriptor: | 2''-aminoglycoside nucleotidyltransferase, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Cox, G, Stogios, P.J, Savchenko, A, Wright, G.D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and Molecular Basis for Resistance to Aminoglycoside Antibiotics by the Adenylyltransferase ANT(2)-Ia.
Mbio, 6, 2015
|
|
8SPM
 
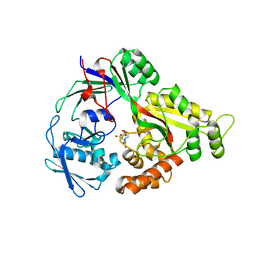 | | Crystal structure of NikA in complex Ni-AMA | | Descriptor: | Aspergillomarasmine A, NICKEL (II) ION, Nickel ABC transporter, ... | | Authors: | Sychantha, D, Prehna, G, Wright, G.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Targeting bacterial nickel transport with aspergillomarasmine A suppresses virulence-associated Ni-dependent enzymes.
Nat Commun, 15, 2024
|
|
8UK6
 
 | |
7UUK
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with tobramycin | | Descriptor: | Aminocyclitol acetyltransferase ApmA, CHLORIDE ION, TOBRAMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUN
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with neomycin | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, NEOMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Osipiuk, J, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUO
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA H135A mutant, complex with tobramycin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, COENZYME A, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUL
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with kanamycin B and coenzyme A | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUM
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with paromomycin and coenzyme A | | Descriptor: | Aminocyclitol acetyltransferase ApmA, COENZYME A, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
2XDO
 
 | | Structure of the Tetracycline degrading Monooxygenase TetX2 from Bacteroides thetaiotaomicron | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TETX2 PROTEIN | | Authors: | Volkers, G, Palm, G.J, Wright, G.D, Hinrichs, W. | | Deposit date: | 2010-05-05 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
3TYK
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia | | Descriptor: | CHLORIDE ION, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | Authors: | Stogios, P.J, Shabalin, I.G, Shakya, T, Evdokmova, E, Fan, Y, Chruszcz, M, Minor, W, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of APH(4)-Ia, a hygromycin B resistance enzyme.
J.Biol.Chem., 286, 2011
|
|
2Y6R
 
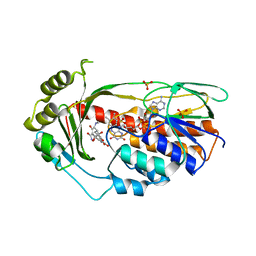 | | Structure of the TetX monooxygenase in complex with the substrate 7- chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Wright, G.D, Hinrichs, W. | | Deposit date: | 2011-01-25 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
2Y6Q
 
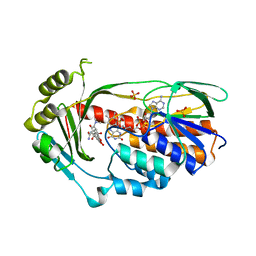 | | Structure of the TetX monooxygenase in complex with the substrate 7- Iodtetracycline | | Descriptor: | 7-IODOTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Wright, G.D, Hinrichs, W. | | Deposit date: | 2011-01-25 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
6BRD
 
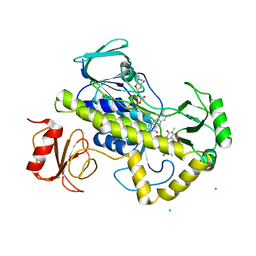 | | Crystal structure of rifampin monooxygenase from Streptomyces venezuelae, complexed with rifampin and FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Cox, G, Kelso, J, Stogios, P.J, Savchenko, A, Anderson, W.F, Wright, G.D, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-30 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action.
Cell Chem Biol, 25, 2018
|
|
4EJ7
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, ATP-bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aminoglycoside 3'-phosphotransferase AphA1-IAB, CALCIUM ION, ... | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Evdokimova, E, Egorova, O, Di Leo, R, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-18 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
5FA7
 
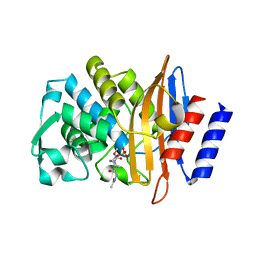 | | CTX-M-15 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-6-(acetamidocarbamoyl)-1-methanoyl-piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
7MK5
 
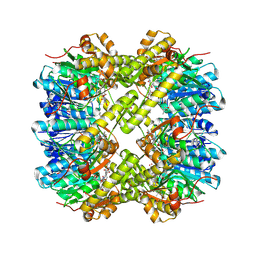 | | Crystal structure of Escherichia coli ClpP covalently inhibited by clipibicyclene | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-[(1E)-3-{[(2E,4E,6E,8S)-8-hydroxy-4-methyldeca-2,4,6-trienoyl]amino}-3-oxoprop-1-en-1-yl]azete-1(2H)-carboxylic acid, ACETATE ION, ... | | Authors: | Culp, E.J, Sychantha, D, Hobson, C, Pawlowski, A.J, Prehna, G, Wright, G.D. | | Deposit date: | 2021-04-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | ClpP inhibitors are produced by a widespread family of bacterial gene clusters.
Nat Microbiol, 7, 2022
|
|
4FEV
 
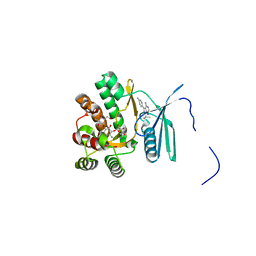 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor pyrazolopyrimidine PP1 | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4GKH
 
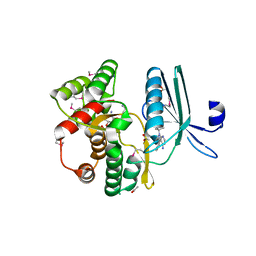 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NA-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
5FAT
 
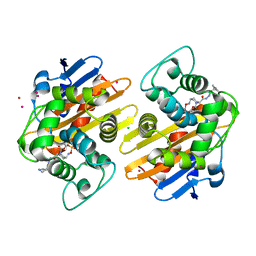 | | OXA-48 in complex with FPI-1602 | | Descriptor: | Beta-lactamase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-12 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
4FEX
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor tyrphostin AG1478 | | Descriptor: | ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, KANAMYCIN A, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
5FAO
 
 | | CTX-M-15 in complex with FPI-1465 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-1-methanoyl-6-[[(3~{S})-pyrrolidin-3-yl]oxycarbamoyl]piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FAS
 
 | | OXA-48 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
4FEU
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor anthrapyrazolone SP600125 | | Descriptor: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
