7KMD
 
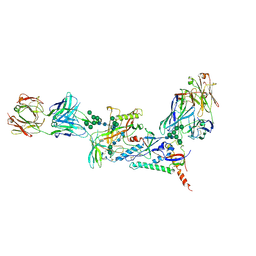 | |
7KLC
 
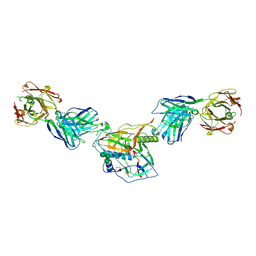 | |
6KVA
 
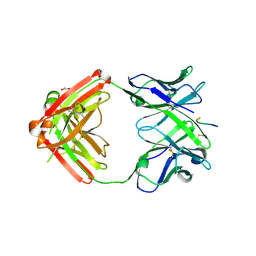 | | Structure of anti-hCXCR2 abN48-2 in complex with its CXCR2 epitope | | Descriptor: | 1,2-ETHANEDIOL, Peptide from C-X-C chemokine receptor type 2, heavy chain, ... | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
6KZ0
 
 | | HRV14 3C in complex with single chain antibody GGVV | | Descriptor: | GGVV H chain, GGVV L chain, Genome polyprotein | | Authors: | Meng, B, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-21 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
25C8
 
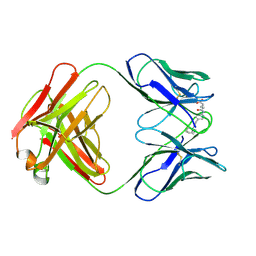 | | CATALYTIC ANTIBODY 5C8, FAB-HAPTEN COMPLEX | | Descriptor: | IGG 5C8, N-METHYL-N-(PARA-GLUTARAMIDOPHENYL-ETHYL)-PIPERIDINIUM ION | | Authors: | Gruber, K, Wilson, I.A. | | Deposit date: | 1998-03-18 | | Release date: | 1999-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antibody catalysis of a disfavored ring closure reaction.
Biochemistry, 38, 1999
|
|
6KVF
 
 | | Structure of anti-hCXCR2 abN48 in complex with its CXCR2 epitope | | Descriptor: | Peptide from C-X-C chemokine receptor type 2, heavy chain, light chain | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
6KYZ
 
 | | HRV14 3C in complex with single chain antibody YDF | | Descriptor: | Genome polyprotein, YDF H chain, YDF L chain | | Authors: | Meng, B, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ME1
 
 | |
6MG4
 
 | | Structure of full-length human lambda-6A light chain JTO | | Descriptor: | JTO light chain | | Authors: | Morgan, G.J, Yan, N.L, Mortenson, D.E, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2018-09-12 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stabilization of amyloidogenic immunoglobulin light chains by small molecules.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MDT
 
 | | Crystal structure of the B41 SOSIP.664 Env trimer with PGT124 and 35O22 Fabs, in P63 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.816 Å) | | Cite: | Capturing the inherent structural dynamics of the HIV-1 envelope glycoprotein fusion peptide.
Nat Commun, 10, 2019
|
|
6M87
 
 | | Fab 10A6 in complex with MPTS | | Descriptor: | 8-methoxypyrene-1,3,6-trisulfonic acid, Fab 10A6 heavy chain, Fab 10A6 light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-08-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure and Dynamics of Stacking Interactions in an Antibody Binding Site.
Biochemistry, 58, 2019
|
|
6MSY
 
 | | Anti-HIV-1 Fab Fab 2G12 + Man4 re-refinement | | Descriptor: | ACETATE ION, Fab 2G12, light chain, ... | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-10-18 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
6MCO
 
 | | Crystal structure of the B41 SOSIP.664 Env trimer with PGT124 and 35O22 Fabs, in P23 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-08-31 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Capturing the inherent structural dynamics of the HIV-1 envelope glycoprotein fusion peptide.
Nat Commun, 10, 2019
|
|
6MG5
 
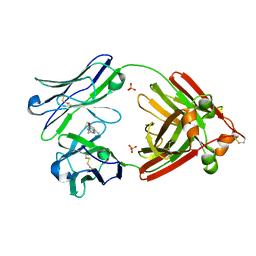 | | Structure of full-length human lambda-6A light chain JTO in complex with coumarin 1 | | Descriptor: | 7-(diethylamino)-4-methyl-2H-1-benzopyran-2-one, Light chain JTO, PHOSPHATE ION | | Authors: | Morgan, G.J, Yan, N.L, Mortenson, D.E, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2018-09-12 | | Release date: | 2019-04-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilization of amyloidogenic immunoglobulin light chains by small molecules.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1ZHN
 
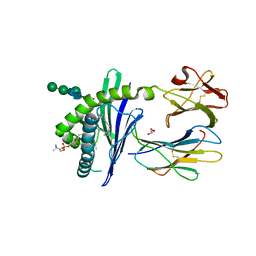 | | Crystal Structure of mouse CD1d bound to the self ligand phosphatidylcholine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(DODECANOYLOXY)METHYL]-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-3,5,8-TRIOXA-4-PHOSPHADOTRIACONTAN-1-AMINIUM 4-OXIDE, CD1d1 antigen, ... | | Authors: | Giabbai, B, Sidobre, S, Crispin, M.M.D, Sanchez Ruiz, Y, Bachi, A, Kronenberg, M, Wilson, I.A, Degano, M. | | Deposit date: | 2005-04-26 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation
J.Immunol., 175, 2005
|
|
6N2X
 
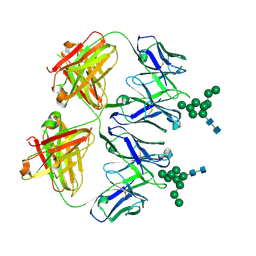 | | Anti-HIV-1 Fab 2G12 + Man9 re-refinement | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Antibody domain exchange is an immunological solution to carbohydrate cluster recognition.
Science, 300, 2003
|
|
6MNF
 
 | | Anti-HIV-1 Fab 2G12 + Man8 re-refinement | | Descriptor: | Fab 2G12, light chain, Fab 2g12, ... | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
6N35
 
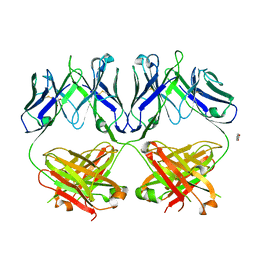 | | Anti-HIV-1 Fab 2G12 + Man1-2 re-refinement | | Descriptor: | BENZOIC ACID, Fab 2G12 heavy chain, Fab 2G12 light chain, ... | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Antibody domain exchange is an immunological solution to carbohydrate cluster recognition.
Science, 300, 2003
|
|
2CKB
 
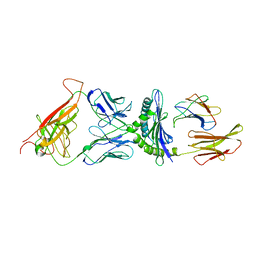 | | STRUCTURE OF THE 2C/KB/DEV8 COMPLEX | | Descriptor: | ALPHA, BETA T CELL RECEPTOR, BETA-2 MICROGLOBULIN, ... | | Authors: | Garcia, K.C, Degano, M, Wilson, I.A. | | Deposit date: | 1998-01-14 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen.
Science, 279, 1998
|
|
7SJS
 
 | |
7SBU
 
 | |
3MJ7
 
 | | Crystal structure of the complex of JAML and Coxsackie and Adenovirus receptor, CAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coxsackievirus and adenovirus receptor homolog, ... | | Authors: | Verdino, P, Wilson, I.A. | | Deposit date: | 2010-04-12 | | Release date: | 2010-09-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The molecular interaction of CAR and JAML recruits the central cell signal transducer PI3K.
Science, 329, 2010
|
|
3MJ6
 
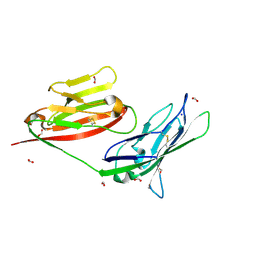 | |
8U08
 
 | |
8V06
 
 | | Crystal structure of mouse PLD3 co-crystallized with 5'Pi-ssDNA for 9 days | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural and mechanistic insights into disease-associated endolysosomal exonucleases PLD3 and PLD4.
Structure, 32, 2024
|
|
