3DWF
 
 | | Crystal Structure of the Guinea Pig 11beta-Hydroxysteroid Dehydrogenase Type 1 Mutant F278E | | Descriptor: | 11-beta-hydroxysteroid dehydrogenase 1, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lawson, A.J, Ride, J.P, White, S.A, Walker, E.A. | | Deposit date: | 2008-07-22 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutations of key hydrophobic surface residues of 11 beta-hydroxysteroid dehydrogenase type 1 increase solubility and monodispersity in a bacterial expression system
Protein Sci., 18, 2009
|
|
7NIY
 
 | | E. coli NfsA with FMN | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-14 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NB9
 
 | | E. coli NfsA with nitrofurantoin | | Descriptor: | 1-[(~{E})-(5-nitrofuran-2-yl)methylideneamino]imidazolidine-2,4-dione, DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.D, Jarrom, D, Grainger, A.I, Parr, R.J, Hyde, E.I, White, S.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NNX
 
 | | E. coli NfsA with 1,4-benzoquinone | | Descriptor: | 1,2-ETHANEDIOL, 1,4-benzoquinone, CHLORIDE ION, ... | | Authors: | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NMP
 
 | | E. coli NfsA with hydroquinone | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Day, M.D, Jarrom, D, Parr, R.J, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
2N5G
 
 | | NMR structure of KorA, a plasmid-encoded, global transcription regulator KorA | | Descriptor: | TrfB transcriptional repressor protein | | Authors: | Rajasekar, K.V, Lovering, A.L, Dancea, F.V, Scott, D.J, Harris, S, Bingle, L.E, Roessle, M, Thomas, C.M, Hyde, E.I, White, S.A. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
1YLU
 
 | | The structure of E. coli nitroreductase with bound acetate, crystal form 2 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
1YKI
 
 | | The structure of E. coli nitroreductase bound with the antibiotic nitrofurazone | | Descriptor: | CITRIC ACID, DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
1YLR
 
 | | The structure of E.coli nitroreductase with bound acetate, crystal form 1 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
2FR8
 
 | | Structure of transhydrogenase (dI.R127A.NAD+)2(dIII.NADP+)1 asymmetric complex | | Descriptor: | NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Brondijk, T.H, van Boxel, G.I, Mather, O.C, Quirk, P.G, White, S.A, Jackson, J.B. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Role of Invariant Amino Acid Residues at the Hydride Transfer Site of Proton-translocating Transhydrogenase.
J.Biol.Chem., 281, 2006
|
|
2FRD
 
 | | Structure of Transhydrogenase (dI.S138A.NADH)2(dIII.NADPH)1 asymmetric complex | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Brondijk, T.H, van Boxel, G.I, Mather, O.C, Quirk, P.G, White, S.A, Jackson, J.B. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Role of Invariant Amino Acid Residues at the Hydride Transfer Site of Proton-translocating Transhydrogenase.
J.Biol.Chem., 281, 2006
|
|
2FSV
 
 | | Structure of transhydrogenase (dI.D135N.NAD+)2(dIII.E155W.NADP+)1 asymmetric complex | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Brondijk, T.H, van Boxel, G.I, Mather, O.C, Quirk, P.G, White, S.A, Jackson, J.B. | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Role of Invariant Amino Acid Residues at the Hydride Transfer Site of Proton-translocating Transhydrogenase.
J.Biol.Chem., 281, 2006
|
|
1PT9
 
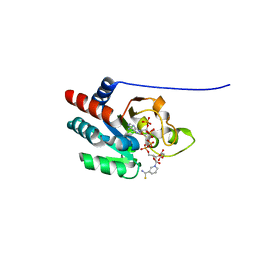 | | Crystal Structure Analysis of the DIII Component of Transhydrogenase with a Thio-Nicotinamide Nucleotide Analogue | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, GLYCEROL, NAD(P) transhydrogenase, ... | | Authors: | Singh, A, Venning, J.D, Quirk, P.G, van Boxel, G.I, Rodrigues, D.J, White, S.A, Jackson, J.B. | | Deposit date: | 2003-06-23 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Interactions between transhydrogenase and thio-nicotinamide analogues of NAD(H) and NADP(H) underline the importance of nucleotide conformational changes in coupling to proton translocation
J.Biol.Chem., 278, 2003
|
|
1NM5
 
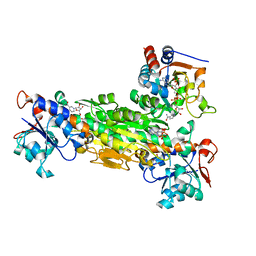 | | R. rubrum transhydrogenase (dI.Q132N)2(dIII)1 asymmetric complex | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Van Boxel, G.I, Quirk, P.G, Cotton, N.P, White, S.A, Jackson, J.B. | | Deposit date: | 2003-01-09 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glutamine 132 in the NAD(H)-binding component of proton-translocating transhydrogenase tethers the nucleotides before hydride transfer.
Biochemistry, 42, 2003
|
|
1S1P
 
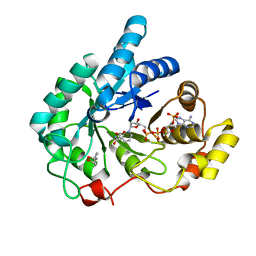 | | Crystal structures of prostaglandin D2 11-ketoreductase (AKR1C3) in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Aldo-keto reductase family 1 member C3, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-07 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
1U2D
 
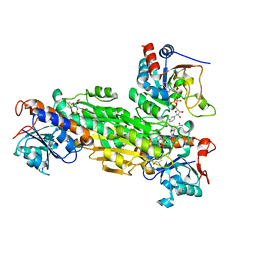 | | Structre of transhydrogenaes (dI.NADH)2(dIII.NADPH)1 asymmetric complex | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-19 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
1S1R
 
 | | Crystal structures of prostaglandin D2 11-ketoreductase (AKR1C3) in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Aldo-keto reductase family 1 member C3, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-07 | | Release date: | 2004-03-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
1S2A
 
 | | Crystal structures of prostaglandin D2 11-ketoreductase in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | Aldo-keto reductase family 1 member C3, DIMETHYL SULFOXIDE, INDOMETHACIN, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
1U2G
 
 | | transhydrogenase (dI.ADPr)2(dIII.NADPH)1 asymmetric complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-19 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
1U28
 
 | | R. rubrum transhydrogenase asymmetric complex (dI.NAD+)2(dIII.NADP+)1 | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-16 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
1S2C
 
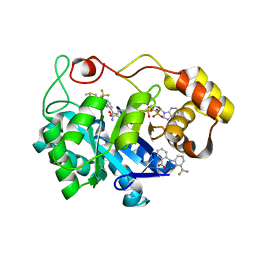 | | Crystal structures of prostaglandin D2 11-ketoreductase in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Aldo-keto reductase family 1 member C3, DIMETHYL SULFOXIDE, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
4QMD
 
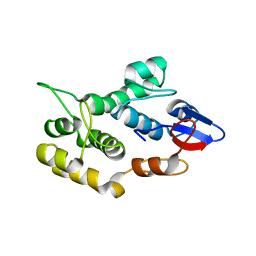 | | Crystal structure of human envoplakin plakin repeat domain | | Descriptor: | Envoplakin | | Authors: | Mohammed, F, Al-Jassar, C, White, S.A, Fogl, C, Jeeves, M, Knowles, T.J, Odinstova, E, Rodriguez-Zamora, P, Overduin, M, Chidgey, M. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Mechanism of intermediate filament recognition by plakin repeat domains revealed by envoplakin targeting of vimentin.
Nat Commun, 7, 2016
|
|
1U31
 
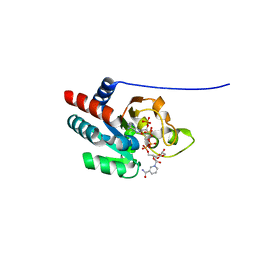 | | recombinant human heart transhydrogenase dIII bound with NADPH | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase, mitochondrial, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-20 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
1NMU
 
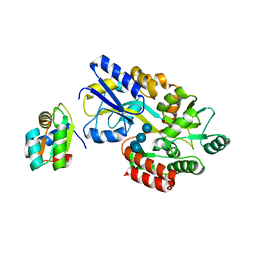 | | MBP-L30 | | Descriptor: | 60S ribosomal protein L30, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding periplasmic protein | | Authors: | Chao, J.A, Prasad, G.S, White, S.A, Stout, C.D, Williamson, J.R. | | Deposit date: | 2003-01-10 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Inherent Protein Structural Flexibility at the RNA-binding Interface of L30e
J.Mol.Biol., 326, 2003
|
|
1PTJ
 
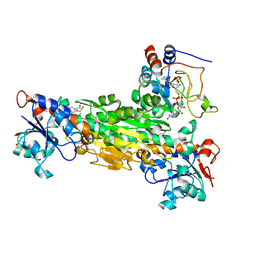 | | Crystal structure analysis of the DI and DIII complex of transhydrogenase with a thio-nicotinamide nucleotide analogue | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Singh, A, Venning, J.D, Quirk, P.G, van Boxel, G.I, Rodrigues, D.J, White, S.A, Jackson, J.B. | | Deposit date: | 2003-06-23 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Interactions between transhydrogenase and thio-nicotinamide analogues of NAD(H) and NADP(H) underline the importance of nucleotide conformational changes in coupling to proton translocation
J.Biol.Chem., 278, 2003
|
|
