1OWJ
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-(1-ISOPROPYL-3,4-DIHYDROISOQUINOLIN-7-YL)-2-NAPHTHAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
1OWH
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-[4-(AMINOMETHYL)PHENYL]-2-NAPHTHAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
1SQT
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 7-METHOXY-8-[1-(METHYLSULFONYL)-1H-PYRAZOL-4-YL]NAPHTHALENE-2-CARBOXIMIDAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhang, X, Mantei, R, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-19 | | Release date: | 2004-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1SQO
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 8-(PYRIMIDIN-2-YLAMINO)NAPHTHALENE-2-CARBOXIMIDAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhao, X, Mantei, R, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-19 | | Release date: | 2004-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2VK1
 
 | | Crystal structure of the Saccharomyces cerevisiae pyruvate decarboxylase variant D28A in complex with its substrate | | Descriptor: | MAGNESIUM ION, PYRUVATE DECARBOXYLASE ISOZYME 1, PYRUVIC ACID, ... | | Authors: | Kutter, S, Weik, M, Weiss, M.S, Konig, S. | | Deposit date: | 2007-12-16 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Covalently Bound Substrate at the Regulatory Site of Yeast Pyruvate Decarboxylases Triggers Allosteric Enzyme Activation.
J.Biol.Chem., 284, 2009
|
|
1SQA
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-[4-(AMINOMETHYL)PHENYL]-4-(PYRIMIDIN-2-YLAMINO)-2-NAPHTHAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhao, X, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-18 | | Release date: | 2004-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
4QLP
 
 | | Atomic structure of tuberculosis necrotizing toxin (TNT) complexed with its immunity factor IFT | | Descriptor: | Alanine and proline rich protein, tuberculosis necrotizing toxin (TNT), immunity factor IFT | | Authors: | Cingolani, G, Lokareddy, R.K, Sun, J, Siroy, A, Speer, A, Doornbos, K.S, Niederweis, M. | | Deposit date: | 2014-06-12 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The tuberculosis necrotizing toxin kills macrophages by hydrolyzing NAD.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2KTT
 
 | | Solution Structure of a Covalently Bound Pyrrolo[2,1-c][1,4]benzodiazepine-Benzimidazole Hybrid to a 10mer DNA Duplex | | Descriptor: | (11aS)-7-methoxy-8-(3-{4-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]phenoxy}propoxy)-1,2,3,10,11,11a-hexahydro-5H-pyrrolo[2,1-c][1,4]benzodiazepin-5-one, 5'-D(*AP*AP*CP*AP*AP*TP*TP*GP*TP*T)-3' | | Authors: | Rettig, M, Weingarth, M, Langel, W, Kamal, A, Kumar, P.P, Weisz, K. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a covalently bound pyrrolo[2,1-c][1,4]benzodiazepine-benzimidazole hybrid to a 10mer DNA duplex.
Biochemistry, 48, 2009
|
|
1YJ5
 
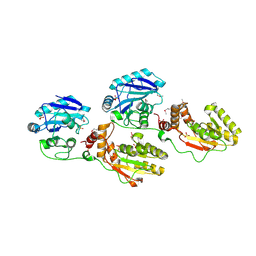 | | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme | | Descriptor: | 5' polynucleotide kinase-3' phosphatase FHA domain, 5' polynucleotide kinase-3' phosphatase catalytic domain, SULFATE ION | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-13 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
1YJM
 
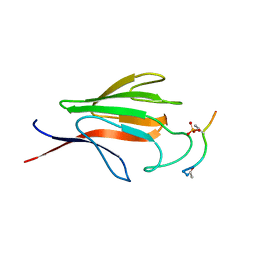 | | Crystal structure of the FHA domain of mouse polynucleotide kinase in complex with an XRCC4-derived phosphopeptide. | | Descriptor: | 12-mer peptide from DNA-repair protein XRCC4, Polynucleotide 5'-hydroxyl-kinase | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
2C5G
 
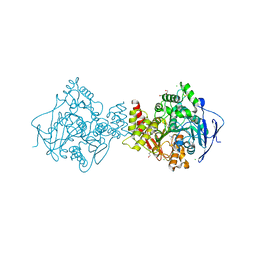 | | Torpedo californica acetylcholinesterase in complex with 20mM thiocholine | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.P, Fournier, D, Greenblatt, H.M, Sussman, J.L, Zaccai, G, Silman, I, Weik, M. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights Into Substrate Traffic and Inhibition in Acetylcholinesterase.
Embo J., 25, 2006
|
|
2C4H
 
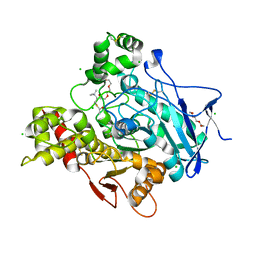 | | Torpedo californica acetylcholinesterase in complex with 500mM acetylthiocholine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ACETYL GROUP, ... | | Authors: | Colletier, J.P, Fournier, D, Greenblatt, H.M, Sussman, J.L, Zaccai, G, Silman, I, Weik, M. | | Deposit date: | 2005-10-19 | | Release date: | 2006-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into substrate traffic and inhibition in acetylcholinesterase.
EMBO J., 25, 2006
|
|
2C5F
 
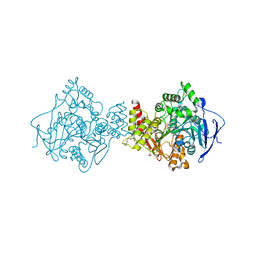 | | Torpedo californica acetylcholinesterase in complex with a non hydrolysable substrate analogue, 4-oxo-N,N,N-trimethylpentanaminium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.P, Fournier, D, Greenblatt, H.M, Sussman, J.L, Zaccai, G, Silman, I, Weik, M. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Substrate Traffic and Inhibition in Acetylcholinesterase.
Embo J., 25, 2006
|
|
2C58
 
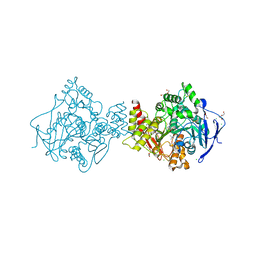 | | Torpedo californica acetylcholinesterase in complex with 20mM acetylthiocholine | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.P, Fournier, D, Greenblatt, H.M, Sussman, J.L, Zaccai, G, Silman, I, Weik, M. | | Deposit date: | 2005-10-26 | | Release date: | 2006-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into Substrate Traffic and Inhibition in Acetylcholinesterase.
Embo J., 25, 2006
|
|
7KNG
 
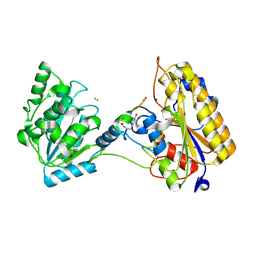 | | 2.10A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-2 Y7F) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, DTY-ASP-TYR-PRO-GLY-ASP-PHE-CYS-TYR-LEU-TYR-GLY-THR-CYS, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
7KNF
 
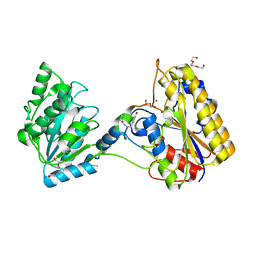 | | 1.80A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-1 NHOH) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, DTY-ASP-TYR-PRO-GLY-ASP-HIS-CYS-TYR-LEU-TYR-GLY-THR, SODIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
2CEK
 
 | | Conformational Flexibility in the Peripheral Site of Torpedo californica Acetylcholinesterase Revealed by the Complex Structure with a Bifunctional Inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sanson, B, Colletier, J.P, Nachon, F, Gabellieri, E, Fattorusso, C, Campiani, G, Weik, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational flexibility in the peripheral site of Torpedo californica acetylcholinesterase revealed by the complex structure with a bifunctional inhibitor.
J. Am. Chem. Soc., 128, 2006
|
|
4A16
 
 | | Structure of mouse Acetylcholinesterase complex with Huprine derivative | | Descriptor: | (1-{4-[(7S,11S)-12-AMINO-3-CHLORO-6,7,10,11-TETRAHYDRO-7,11-METHANOCYCLOOCTA[B]QUINOLIN-9-YL]BUTYL}-1H-1,2,3-TRIAZOL-4-YL)METHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Carletti, E, Colletier, J.P, Nachon, F, Weik, M, Ronco, C, Jean, L, Renard, P.Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Huprine Derivatives as Sub-Nanomolar Human Acetylcholinesterase Inhibitors: From Rational Design to Validation by X-Ray Crystallography.
Chemmedchem, 7, 2012
|
|
4BC1
 
 | | Structure of mouse acetylcholinesterase inhibited by CBDP (30-min soak): cresyl-saligenin-phosphoserine adduct | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, CHLORIDE ION, ... | | Authors: | Carletti, E, Colletier, J.-P, Schopfer, L.M, Santoni, G, Masson, P, Lockridge, O, Nachon, F, Weik, M. | | Deposit date: | 2012-09-30 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Inhibition Pathways of the Potent Organophosphate Cbdp with Cholinesterases Revealed by X-Ray Crystallographic Snapshots and Mass Spectrometry
Chem.Res.Toxicol., 26, 2013
|
|
4BBZ
 
 | | Structure of human butyrylcholinesterase inhibited by CBDP (2-min soak): Cresyl-phosphoserine adduct | | Descriptor: | (2-methylphenyl) dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Carletti, E, Colletier, J.-P, Schopfer, L.M, Santoni, G, Masson, P, Lockridge, O, Nachon, F, Weik, M. | | Deposit date: | 2012-09-30 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition Pathways of the Potent Organophosphate Cbdp with Cholinesterases Revealed by X-Ray Crystallographic Snapshots and Mass Spectrometry
Chem.Res.Toxicol., 26, 2013
|
|
4BC0
 
 | | Structure of mouse acetylcholinesterase inhibited by CBDP (12-h soak) : Cresyl-phosphoserine adduct | | Descriptor: | (2-methylphenyl) dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Carletti, E, Colletier, J.-P, Schopfer, L.M, Santoni, G, Masson, P, Lockridge, O, Nachon, F, Weik, M. | | Deposit date: | 2012-09-30 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Inhibition Pathways of the Potent Organophosphate Cbdp with Cholinesterases Revealed by X-Ray Crystallographic Snapshots and Mass Spectrometry
Chem.Res.Toxicol., 26, 2013
|
|
3LYQ
 
 | | Crystal structure of IpgB2 from Shigella flexneri | | Descriptor: | CITRATE ANION, IpgB2, MU-OXO-DIIRON | | Authors: | Klink, B.U, Barden, S, Heidler, T.V, Borchers, C, Ladwein, M, Stradal, T.E.B, Rottner, K, Heinz, D.W. | | Deposit date: | 2010-02-28 | | Release date: | 2010-03-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Shigella IPGB2 in complex with human RhoA: Implications for the mechanism of bacterial GEF-mimicry
J.Biol.Chem., 285, 2010
|
|
5NX2
 
 | | Crystal structure of thermostabilised full-length GLP-1R in complex with a truncated peptide agonist at 3.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon-like peptide 1 receptor, ... | | Authors: | Rappas, M, Jazayeri, A, Brown, A.J.H, Kean, J, Errey, J.C, Robertson, N, Fiez-Vandal, C, Andrews, S.P, Congreve, M, Bortolato, A, Mason, J.S, Baig, A.H, Teobald, I, Dore, A.S, Weir, M, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2017-05-09 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of the GLP-1 receptor bound to a peptide agonist.
Nature, 546, 2017
|
|
2VA9
 
 | | Structure of native TcAChE after a 9 seconds annealing to room temperature during the first 5 seconds of which laser irradiation at 266nm took place | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE | | Authors: | Colletier, J.-P, Sanson, B, Royant, A, Specht, A, Nachon, F, Masson, P, Zaccai, G, Sussman, J.L, Goeldner, M, Silman, I, Bourgeois, D, Weik, M. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Use of a 'Caged' Analog to Study Traffic of Choline within Acetylcholinesterase by Kinetic Crystallography
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2V97
 
 | | Structure of the unphotolysed complex of TcAChE with 1-(2- nitrophenyl)-2,2,2-trifluoroethyl-arsenocholine after a 9 seconds annealing to room temperature | | Descriptor: | 1-(2-nitrophenyl)-2,2,2-trifluoroethyl]-arsenocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.-P, Sanson, B, Royant, A, Specht, A, Nachon, F, Masson, P, Zaccai, G, Sussman, J.L, Goeldner, M, Silman, I, Bourgeois, D, Weik, M. | | Deposit date: | 2007-08-22 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Use of a 'Caged' Analog to Study Traffic of Choline within Acetylcholinesterase by Kinetic Crystallography
Acta Crystallogr.,Sect.D, 63, 2007
|
|
