2VG9
 
 | | Crystal structure of Loop Swap mutant of Necallimastix patriciarum Xyn11A | | Descriptor: | ACETATE ION, BIFUNCTIONAL ENDO-1,4-BETA-XYLANASE A, CADMIUM ION | | Authors: | Vardakou, M, Dumon, C, Flint, J.E, Murray, J.W, Christakopoulos, P, Weiner, D.P, Juge, N, Lewis, R.J, Gilbert, H.J. | | Deposit date: | 2007-11-09 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the Structural Basis for Substrate and Inhibitor Recognition in Eukaryotic Gh11 Xylanases.
J.Mol.Biol., 375, 2008
|
|
5N6L
 
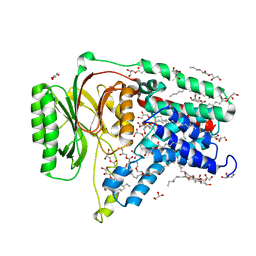 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt C387A mutant from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
5N6H
 
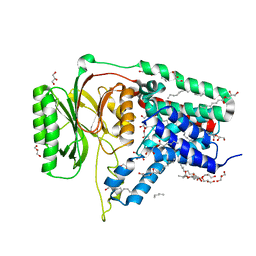 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
8PKJ
 
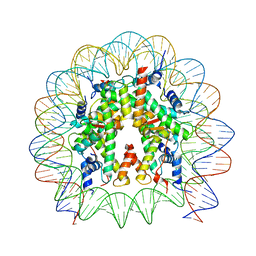 | | Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PKI
 
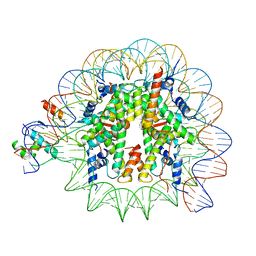 | | Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B type 1-C/E/G, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5N6M
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
5C7X
 
 | | Crystal structure of MOR04357, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, DI(HYDROXYETHYL)ETHER, Granulocyte-macrophage colony-stimulating factor, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
8BQ2
 
 | | CryoEM structure of the pre-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Appleby, R, Bollschweiler, D, Pellegrini, L. | | Deposit date: | 2022-11-18 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly.
Iscience, 26, 2023
|
|
5D70
 
 | | Crystal structure of MOR03929, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, heavy chain, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-13 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
5D7S
 
 | | Crystal structure of MOR04357, a neutralizing anti-human GM-CSF antibody Fab fragment | | Descriptor: | (2S,3S)-butane-2,3-diol, Immunglobulin G1 Fab fragment, heavy chain, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-10-14 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
8OJ2
 
 | |
5D71
 
 | | Crystal structure of MOR04302, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | DI(HYDROXYETHYL)ETHER, Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-13 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
8OJ1
 
 | |
5D72
 
 | | Crystal structure of MOR04252, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | DI(HYDROXYETHYL)ETHER, Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-13 | | Release date: | 2015-10-14 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
8BR2
 
 | | CryoEM structure of the post-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*GP*CP*GP*AP*GP*CP*TP*CP*GP*AP*TP*GP*CP*AP*CP*CP*TP*CP*CP*A)-3'), ... | | Authors: | Appleby, R, Bollschweiler, D, Pellegrini, L. | | Deposit date: | 2022-11-22 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly.
Iscience, 26, 2023
|
|
3RV1
 
 | | Crystal structure of the N-terminal and RNase III domains of K. polysporus Dcr1 E224Q mutant | | Descriptor: | K. polysporus Dcr1 | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2011-05-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | The inside-out mechanism of dicers from budding yeasts.
Cell(Cambridge,Mass.), 146, 2011
|
|
3RV0
 
 | | Crystal structure of K. polysporus Dcr1 without the C-terminal dsRBD | | Descriptor: | K. polysporus Dcr1, MAGNESIUM ION | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2011-05-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The inside-out mechanism of dicers from budding yeasts.
Cell(Cambridge,Mass.), 146, 2011
|
|
6YCQ
 
 | | Crystal structure of the DNA binding domain of Arabidopsis thaliana Auxin Response Factor 1 (AtARF1) in complex with High Affinity DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 21-7A, 21-7B, ... | | Authors: | Crespo, I, Weijers, D, Boer, D.R. | | Deposit date: | 2020-03-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2VGD
 
 | | Crystal structure of environmental isolated GH11 in complex with xylobiose and feruloyl-arabino-xylotriose | | Descriptor: | 5-O-[(2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoyl]-alpha-L-ribofuranose, ENXYN11A, GLYCEROL, ... | | Authors: | Vardakou, M, Dumon, C, Flint, J.E, Murray, J.W, Christakopoulos, P, Weiner, D.P, Juge, N, Lewis, R.J, Gilbert, H.J. | | Deposit date: | 2007-11-12 | | Release date: | 2007-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Understanding the Structural Basis for Substrate and Inhibitor Recognition in Eukaryotic Gh11 Xylanases.
J.Mol.Biol., 375, 2008
|
|
4LDU
 
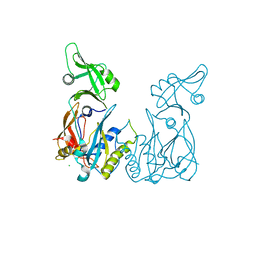 | | Crystal structure of the DNA binding domain of Arabidopsis thaliana auxin response factor 5 | | Descriptor: | Auxin response factor 5, CHLORIDE ION | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
1KKD
 
 | | Solution structure of the calmodulin binding domain (CaMBD) of small conductance Ca2+-activated potassium channels (SK2) | | Descriptor: | Small conductance calcium-activated potassium channel protein 2 | | Authors: | Wissmann, R, Bildl, W, Neumann, H, Rivard, A.F, Kloecker, N, Weitz, D, Schulte, U, Adelman, J.P, Bentrop, D, Fakler, B. | | Deposit date: | 2001-12-07 | | Release date: | 2001-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A helical region in the C terminus of small-conductance Ca2+-activated K+ channels controls assembly with apo-calmodulin.
J.Biol.Chem., 277, 2002
|
|
4LDY
 
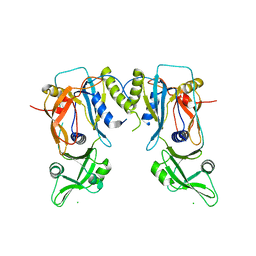 | | Crystal structure of the DNA binding domain of the G245A mutant of arabidopsis thaliana auxin reponse factor 1 | | Descriptor: | Auxin response factor 1, CHLORIDE ION | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
4LDV
 
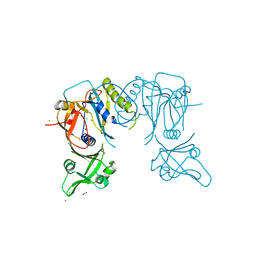 | | Crystal structure of the DNA binding domain of A. thailana auxin response factor 1 | | Descriptor: | Auxin response factor 1, CHLORIDE ION, FORMIC ACID, ... | | Authors: | boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
6SDG
 
 | |
1CD0
 
 | | STRUCTURE OF HUMAN LAMDA-6 LIGHT CHAIN DIMER JTO | | Descriptor: | PROTEIN (JTO, A VARIABLE DOMAIN FROM LAMBDA-6 TYPE IMMUNOGLOBULIN LIGHT CHAIN) | | Authors: | Pokkuluri, P.R, Solomon, A, Weiss, D.T, Stevens, F.J, Schiffer, M. | | Deposit date: | 1999-03-05 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tertiary structure of human lambda 6 light chains.
Amyloid, 6, 1999
|
|
