3QK5
 
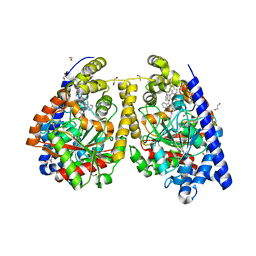 | | Crystal structure of fatty acid amide hydrolase with small molecule inhibitor | | Descriptor: | (3-{(3R)-1-[4-(1-benzothiophen-2-yl)pyrimidin-2-yl]piperidin-3-yl}-2-methyl-1H-pyrrolo[2,3-b]pyridin-1-yl)acetonitrile, 1,2-ETHANEDIOL, Fatty-acid amide hydrolase 1, ... | | Authors: | Min, X, Walker, N.P.C, Wang, Z. | | Deposit date: | 2011-01-31 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of potent, noncovalent fatty acid amide hydrolase (FAAH) inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3VZC
 
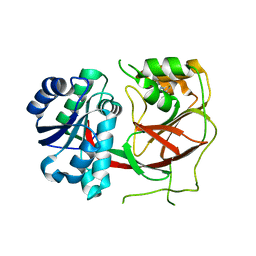 | | Crystal structure of Sphingosine Kinase 1 with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-(4-chlorophenyl)-1,3-thiazol-2-yl]amino}phenol, Sphingosine kinase 1 | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-11 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3QKV
 
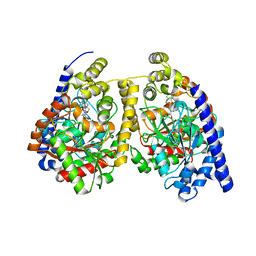 | | Crystal structure of fatty acid amide hydrolase with small molecule compound | | Descriptor: | (6-bromo-1'H,4H-spiro[1,3-benzodioxine-2,4'-piperidin]-1'-yl)methanol, Fatty-acid amide hydrolase 1 | | Authors: | Min, X, Walker, N.P.C, Wang, Z. | | Deposit date: | 2011-02-01 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7CUU
 
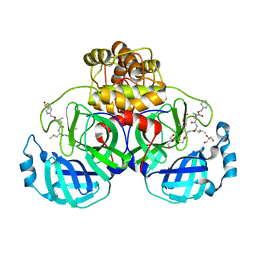 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with MG132 | | Descriptor: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | Deposit date: | 2020-08-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
5C6H
 
 | | Mcl-1 complexed with Mule | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mule BH3 peptide from E3 ubiquitin-protein ligase HUWE1 | | Authors: | Song, T, Wang, Z, Ji, F, Chai, G, Liu, Y, Li, X, Li, Z, Fan, Y, Zhang, Z. | | Deposit date: | 2015-06-23 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Mcl-1 complexed with Mule at 2.05 Angstroms resolution
To Be Published
|
|
6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
7CU5
 
 | | N-Glycosylation of PD-1 and glycosylation dependent binding of PD-1 specific monoclonal antibody camrelizumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Programmed cell death protein 1, ... | | Authors: | Liu, K.F, Tan, S.G, Jin, W.J, Guan, J.W, Wang, W.L, Sun, H, Qi, J.X, Yan, J.H, Chai, Y, Wang, Z.F, Chu, X.D, Gao, G.F. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | N-glycosylation of PD-1 promotes binding of camrelizumab.
Embo Rep., 21, 2020
|
|
8GCE
 
 | | The Extracellular Domain of Integrin AlphaIIbBeta3 in Intermediate State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huo, T, Wu, H, Wang, Z. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Full-length alpha IIb beta 3 cryo-EM structure reveals intact integrin initiate-activation intrinsic architecture.
Structure, 32, 2024
|
|
3QJ9
 
 | | Crystal structure of fatty acid amide hydrolase with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-{(3S)-1-[4-(1-benzofuran-2-yl)pyrimidin-2-yl]piperidin-3-yl}-3-ethyl-1,3-dihydro-2H-benzimidazol-2-one, Fatty-acid amide hydrolase 1, ... | | Authors: | Min, X, Walker, N.P.C, Wang, Z. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3O7R
 
 | | Crystal structure of Ru(p-cymene)/apo-H49AFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Takezawa, Y, Bockmann, P, Sugi, N, Wang, Z, Abe, S, Murakami, T, Hikage, T, Erker, G, Watanabe, Y, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-07-31 | | Release date: | 2011-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Incorporation of organometallic Ru complexes into apo-ferritin cage.
J.CHEM.SOC.,DALTON TRANS., 40, 2011
|
|
6IR2
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM2 at 1.4 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM2 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.393 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
3O7S
 
 | | Crystal structure of Ru(p-cymene)/apo-Fr | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4-(1-methylethyl)benzene, CADMIUM ION, ... | | Authors: | Takezawa, Y, Bockmann, P, Sugi, N, Wang, Z, Abe, S, Murakami, T, Hikage, T, Erker, G, Watanabe, Y, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-07-31 | | Release date: | 2011-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Incorporation of organometallic Ru complexes into apo-ferritin cage.
J.Chem.Soc.,Dalton Trans., 40, 2011
|
|
5UJV
 
 | | Crystal structure of FePYR1 in complex with abscisic acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, PYR1 | | Authors: | Ren, Z, Wang, Z, Zhou, X.E, Hong, Y, Cao, M, Chan, Z, Liu, X, Shi, H, Xu, H.E, Zhu, J.-K. | | Deposit date: | 2017-01-19 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure determination and activity manipulation of the turfgrass ABA receptor FePYR1.
Sci Rep, 7, 2017
|
|
4DN5
 
 | | Crystal Structure of NF-kB-inducing Kinase (NIK) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Min, X, Liu, J, Sudom, A, Walker, N.P, Wang, Z. | | Deposit date: | 2012-02-08 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Nuclear Factor Kappa B-inducing kinase domain reveals a constitutively active conformation
J.Biol.Chem., 287, 2012
|
|
3VZB
 
 | | Crystal structure of Sphingosine Kinase 1 | | Descriptor: | (2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
7CPX
 
 | | Lovastatin nonaketide synthase | | Descriptor: | Lovastatin nonaketide synthase, polyketide synthase component, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2020-08-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for the biosynthesis of lovastatin.
Nat Commun, 12, 2021
|
|
8GCD
 
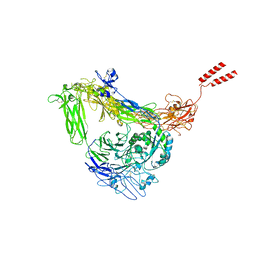 | | Full length Integrin AlphaIIbBeta3 in inactive state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huo, T, Wu, H, Wang, Z. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Full-length alpha IIb beta 3 cryo-EM structure reveals intact integrin initiate-activation intrinsic architecture.
Structure, 32, 2024
|
|
7CPY
 
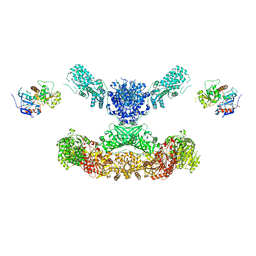 | | Lovastatin nonaketide synthase with LovC | | Descriptor: | Lovastatin nonaketide synthase, enoyl reductase component lovC, polyketide synthase component, ... | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2020-08-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for the biosynthesis of lovastatin.
Nat Commun, 12, 2021
|
|
6UVQ
 
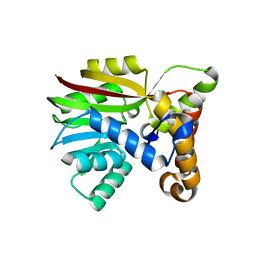 | | Crystal structure of Apo AtmM | | Descriptor: | ACETATE ION, D-glucose O-methyltransferase, MAGNESIUM ION | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of Apo AtmM
To Be Published
|
|
6KTH
 
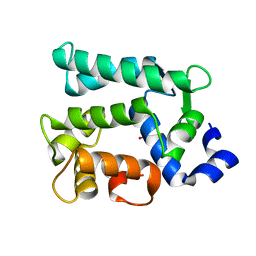 | | Crystal structure of Juvenile hormone diol kinase JHDK-L2 from silkworm, Bombyx mori | | Descriptor: | CALCIUM ION, GLYCEROL, Juvenile hormone diol kinase | | Authors: | Zhang, Y.S, Xu, H.Y, Wang, Z, Zhang, L, Zhao, P, Guo, P.C. | | Deposit date: | 2019-08-28 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural characterization and functional analysis of juvenile hormone diol kinase from the silkworm, Bombyx mori.
Int.J.Biol.Macromol., 167, 2021
|
|
7LUR
 
 | |
5XPV
 
 | | Structure of the V domain of amphioxus IgVJ-C2 | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
7CUT
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with Z-VAD-FMK | | Descriptor: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | Deposit date: | 2020-08-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
5Y6M
 
 | | Zika virus helicase in complex with ADP-AlF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase domain from Genome polyprotein, ... | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
