8EFQ
 
 | | DAMGO-bound mu-opioid receptor-Gi complex | | Descriptor: | DAMGO, ETHANOLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
1KV0
 
 | | Cis/trans Isomerization of Non-prolyl Peptide Bond Observed in Crystal Structure of an Scorpion Toxin | | Descriptor: | Alpha-like toxin BmK-M7 | | Authors: | Guan, R.J, He, X.L, Wang, M, Xiang, Y, Wang, D.C. | | Deposit date: | 2002-01-23 | | Release date: | 2003-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural mechanism governing cis and trans isomeric states and an intramolecular switch for cis/trans isomerization of a non-proline peptide bond observed in crystal structures of scorpion toxins.
J.Mol.Biol., 341, 2004
|
|
4DBF
 
 | | Crystal structures of Cg1458 | | Descriptor: | 2-HYDROXYHEPTA-2,4-DIENE-1,7-DIOATE ISOMERASE, MAGNESIUM ION | | Authors: | Ran, T.T, Xu, D.Q, Wang, W.W, Gao, Y.Y, Wang, M.T. | | Deposit date: | 2012-01-15 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for FAH family.
Biochem.J., 449, 2013
|
|
8ZV9
 
 | | Complex structure of HLA2402 with recognizing SARS-CoV-2 Y453F epitope NYNYLFRLF | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Deng, S.S, Jin, T.C, Xu, Z.H, Wang, M.H. | | Deposit date: | 2024-06-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.
J.Biol.Chem., 300, 2024
|
|
6LN2
 
 | | Crystal structure of full length human GLP1 receptor in complex with Fab fragment (Fab7F38) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab7F38_heavy chain, Fab7F38_light chain, ... | | Authors: | Wu, F, Yang, L, Hang, K, Laursen, M, Wu, L, Han, G.W, Ren, Q, Roed, N.K, Lin, G, Hanson, M, Jiang, H, Wang, M, Reedtz-Runge, S, Song, G, Stevens, R.C. | | Deposit date: | 2019-12-28 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Full-length human GLP-1 receptor structure without orthosteric ligands.
Nat Commun, 11, 2020
|
|
4TN8
 
 | | Crystal structure of Thermus Thermophilus thioredoxin solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4TNO
 
 | | Hypothetical protein PF1117 from Pyrococcus Furiosus: Structure solved by sulfur-SAD using Swiss Light Source Data | | Descriptor: | CHLORIDE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
8Z1E
 
 | | A homotrimeric GPCR architecture of the human cytomegalovirus (UL78) revealed by cryo-EM | | Descriptor: | Uncharacterized protein UL78 | | Authors: | Chen, Y, Li, Y, Zhou, Q, Cong, Z, Lin, S, Yan, J, Chen, X, Yang, D, Ying, T, Wang, M.-W. | | Deposit date: | 2024-04-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | A homotrimeric GPCR architecture of the human cytomegalovirus revealed by cryo-EM.
Cell Discov, 10, 2024
|
|
8YE4
 
 | | The complex of TCR NYN-I and HLA-A24 bound to SARS-CoV-2 Spike448-456 peptide NYNYLYRLF | | Descriptor: | Beta-2-microglobulin, MHC class I antigen precusor, Spike protein S1, ... | | Authors: | Deng, S.S, Jin, T.C, Xu, Z.H, Wang, M.H. | | Deposit date: | 2024-02-21 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.
J.Biol.Chem., 300, 2024
|
|
7BYJ
 
 | | Crystal structure of the FERM domain of FRMPD4 | | Descriptor: | FERM and PDZ domain-containing protein 4 | | Authors: | Lin, L, Wang, M, Wang, C, Zhu, J. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of the FERM domain of a neural scaffold protein FRMPD4 implicated in X-linked intellectual disability.
Biochem.J., 477, 2020
|
|
6LYW
 
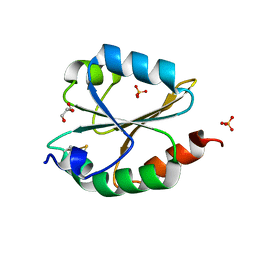 | | Structural insight into the biological functions of Arabidopsis thaliana ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
9JFB
 
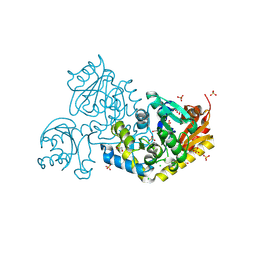 | | Crystal structure of L-threonine-O-3-phosphate decarboxylase CobC | | Descriptor: | CHLORIDE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Jiang, M, Guo, S, Chen, X, Wei, Q, Wang, M. | | Deposit date: | 2024-09-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of l-threonine-O-3-phosphate decarboxylase CobC from Sinorhizobium meliloti involved in vitamin B 12 biosynthesis.
Biochem.Biophys.Res.Commun., 734, 2024
|
|
9FX7
 
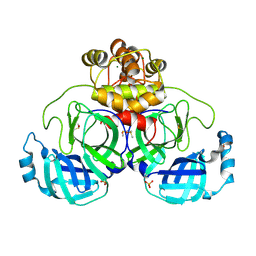 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 294K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9JFF
 
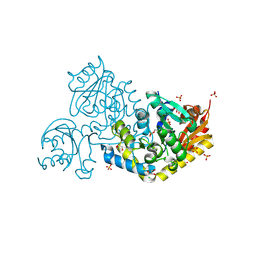 | | Crystal structure of L-threonine-O-3-phosphate decarboxylase CobC in complex with reaction intermediate | | Descriptor: | CHLORIDE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Jiang, M, Guo, S, Chen, X, Wei, Q, Wang, M. | | Deposit date: | 2024-09-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of l-threonine-O-3-phosphate decarboxylase CobC from Sinorhizobium meliloti involved in vitamin B 12 biosynthesis.
Biochem.Biophys.Res.Commun., 734, 2024
|
|
8XXL
 
 | | Cryo-EM structure of the human 40S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
6LYX
 
 | | Crystal structure of oxidized ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Cai, W.G, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
8XXM
 
 | | Cryo-EM structure of the human 40S ribosome with PDCD4 and eIF3G | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8XXN
 
 | | Cryo-EM structure of the human 43S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8IOW
 
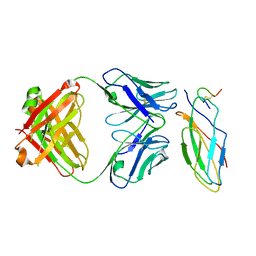 | | Cryo-EM structure of the sarilumab Fab/IL-6R complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Sarilumab Fab, Interleukin-6 receptor subunit alpha, ... | | Authors: | She, J, Chen, L, Wang, M.X. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into IL-6 signaling inhibition by therapeutic antibodies.
Cell Rep, 43, 2024
|
|
8YW5
 
 | | Cryo-EM structure of the retatrutide-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, W.Z, Zhou, Q.T, Cong, Z.T, Yuan, Q.N, Li, W.X, Zhao, F.H, Xu, H.E, Zhao, L.H, Yang, D.H, Wang, M.W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into the triple agonism at GLP-1R, GIPR and GCGR manifested by retatrutide.
Cell Discov, 10, 2024
|
|
4YY0
 
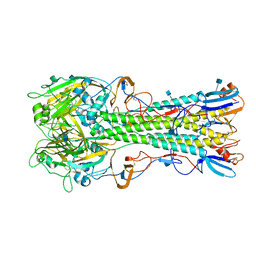 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2 | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
3CFR
 
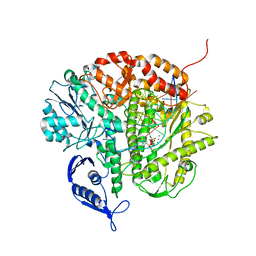 | | Structure of the replicating complex of a POL Alpha family DNA Polymerase, ternary complex 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DGP*DGP*DAP*DCP*DTP*DGP*DCP*DTP*DTP*DAP*(DOC))-3'), ... | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
4WBQ
 
 | | Crystal structure of the exonuclease domain of QIP (QDE-2 interacting protein) solved by native-SAD phasing. | | Descriptor: | CALCIUM ION, QDE-2-interacting protein | | Authors: | Boland, A, Weinert, T, Weichenrieder, O, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
3CFO
 
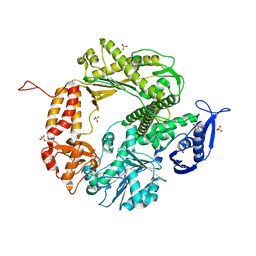 | | Triple Mutant APO structure | | Descriptor: | DNA polymerase, GUANOSINE, SULFATE ION | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
