5AYR
 
 | | The crystal structure of SAUGI/human UDG complex | | Descriptor: | MAGNESIUM ION, Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Huang, M.F, Wang, A.H.J. | | Deposit date: | 2015-09-02 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Using structural-based protein engineering to modulate the differential inhibition effects of SAUGI on human and HSV uracil DNA glycosylase.
Nucleic Acids Res., 44, 2016
|
|
5AYS
 
 | | Crystal structure of SAUGI/HSV UDG complex | | Descriptor: | Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Huang, M.F, Wang, A.H.J. | | Deposit date: | 2015-09-02 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Using structural-based protein engineering to modulate the differential inhibition effects of SAUGI on human and HSV uracil DNA glycosylase.
Nucleic Acids Res., 44, 2016
|
|
8J8Y
 
 | |
3X0T
 
 | | Crystal structure of PirA | | Descriptor: | NITRATE ION, Uncharacterized protein | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J, Lo, C.F. | | Deposit date: | 2014-10-22 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3X0U
 
 | | Crystal structure of PirB | | Descriptor: | Uncharacterized protein | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J, Lo, C.F. | | Deposit date: | 2014-10-22 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3W1O
 
 | | Neisseria DNA mimic protein DMP12 | | Descriptor: | DNA mimic protein DMP12, MAGNESIUM ION | | Authors: | Wang, H.C, Ko, T.P, Wu, M.L, Wang, A.H.J. | | Deposit date: | 2012-11-19 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Neisseria conserved hypothetical protein DMP12 is a DNA mimic that binds to histone-like HU protein
Nucleic Acids Res., 41, 2013
|
|
3WDG
 
 | | Staphylococcus aureus UDG / UGI complex | | Descriptor: | Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor.
Nucleic Acids Res., 42, 2013
|
|
3WDF
 
 | | Staphylococcus aureus UDG | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor.
Nucleic Acids Res., 42, 2013
|
|
6A9S
 
 | | The crystal structure of vaccinia virus A26 (residues 1-397) | | Descriptor: | 1,2-ETHANEDIOL, Protein A26 | | Authors: | Wang, H.C, Ko, T.Z, Luo, Y.C, Liao, Y.T, Chang, W. | | Deposit date: | 2018-07-16 | | Release date: | 2019-06-12 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Vaccinia viral A26 protein is a fusion suppressor of mature virus and triggers membrane fusion through conformational change at low pH.
Plos Pathog., 15, 2019
|
|
2OUV
 
 | | crystal structure of pde10a2 mutant of D564N | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUY
 
 | | crystal structure of pde10a2 mutant D564A in complex with cAMP. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUP
 
 | | crystal structure of PDE10A | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUR
 
 | | crystal structure of PDE10A2 mutant D674A in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUN
 
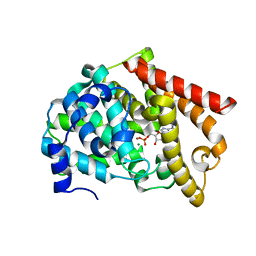 | | crystal structure of PDE10A2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H, Ke, H.M. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUU
 
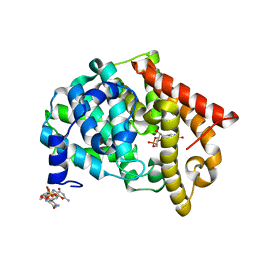 | | crystal structure of PDE10A2 mutant D674A in complex with cGMP | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUS
 
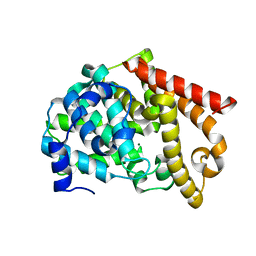 | | crystal structure of PDE10A2 mutant D674A | | Descriptor: | MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUQ
 
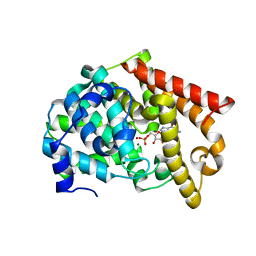 | | crystal structure of PDE10A2 in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6LYJ
 
 | | The crystal structure of SAUGI/EBVUDG complex | | Descriptor: | SAUGI, Uracil-DNA glycosylase | | Authors: | Liao, Y.T, Ko, T.P, Wang, H.C. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into the differential interactions between the DNA mimic protein SAUGI and two gamma herpesvirus uracil-DNA glycosylases.
Int.J.Biol.Macromol., 160, 2020
|
|
6LYV
 
 | | The crystal structure of SAUGI/KSHVUDG complex | | Descriptor: | SAUGI, Uracil-DNA glycosylase | | Authors: | Liao, Y.T, Ko, T.P, Wang, H.C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into the differential interactions between the DNA mimic protein SAUGI and two gamma herpesvirus uracil-DNA glycosylases.
Int.J.Biol.Macromol., 160, 2020
|
|
3WUR
 
 | | Structure of DMP19 Complex with 18-crown-6 | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, L(+)-TARTARIC ACID, ... | | Authors: | Lee, C.C, Wang, H.C, Wang, A.H.J. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8INI
 
 | | vaccinia H2 protein without the transmembrane region | | Descriptor: | TETRAETHYLENE GLYCOL, vaccinia h2 protein | | Authors: | Liu, C.Y, Ko, T.P, Wang, H.C, Chang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional analyses of viral H2 protein of the vaccinia virus entry fusion complex.
J.Virol., 97, 2023
|
|
3WX4
 
 | |
6INR
 
 | | The crystal structure of phytoplasmal effector causing phyllody symptoms 1 (PHYL1) | | Descriptor: | CADMIUM ION, Putative effector, AYWB SAP54-like protein | | Authors: | Liao, Y.T, Lin, S.S, Ko, T.P, Wang, H.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural insights into the interaction between phytoplasmal effector causing phyllody 1 and MADS transcription factors.
Plant J., 100, 2019
|
|
6WOC
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 5-diphosphoinositol pentakisphosphate and Mg, presoaked with 5-IP7, Mg and Fluoride, soaking 1min in the absence of Fluoride. | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
6WO9
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 1-diphosphoinositol pentakisphosphate (1-IP7) and Mg | | Descriptor: | (1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
