6X5I
 
 | |
5XBW
 
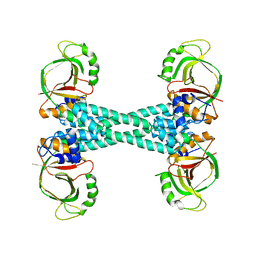 | | The structure of BrlR | | Descriptor: | Probable transcriptional regulator | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
1YM0
 
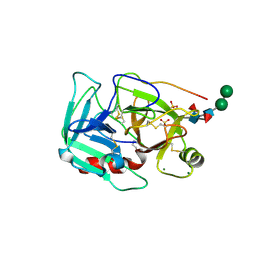 | | Crystal Structure of Earthworm Fibrinolytic Enzyme Component B: a Novel, Glycosylated Two-chained Trypsin | | Descriptor: | MAGNESIUM ION, SULFATE ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, F, Wang, C, Li, M, Zhang, J.P, Gui, L.L, An, X.M, Chang, W.R. | | Deposit date: | 2005-01-20 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of earthworm fibrinolytic enzyme component B: a novel, glycosylated two-chained trypsin.
J.Mol.Biol., 348, 2005
|
|
2Y1R
 
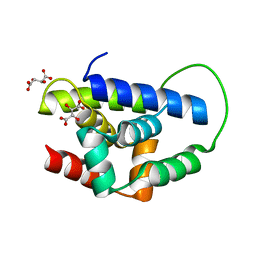 | | Structure of MecA121 & ClpC N-domain complex | | Descriptor: | ADAPTER PROTEIN MECA 1, NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, S,R MESO-TARTARIC ACID | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
2Y1Q
 
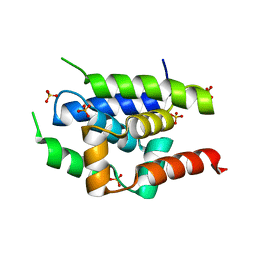 | | Crystal Structure of ClpC N-terminal Domain | | Descriptor: | NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, SULFATE ION | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
7DS7
 
 | | The Crystal Structure of Leaf-branch compost cutinase from Biortus. | | Descriptor: | CITRIC ACID, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Chu, F, Xu, X, Tan, J. | | Deposit date: | 2020-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Leaf-branch compost cutinase from Biortus.
To Be Published
|
|
7DSF
 
 | | The Crystal Structure of human SPR from Biortus. | | Descriptor: | ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sepiapterin reductase, ... | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Meng, Q, Zhang, B, Huang, Y. | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of human SPR from Biortus.
To Be Published
|
|
7D2C
 
 | | The Crystal Structure of human PARP14 from Biortus. | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP14 | | Authors: | Wang, F, Miao, Q, Lv, Z, Cheng, W, Lin, D, Xu, X, Tan, J. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The Crystal Structure of human PARP14 from Biortus.
To Be Published
|
|
7CM2
 
 | | The Crystal Structure of human USP7 USP domain from Biortus | | Descriptor: | GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Wang, F, Cheng, W, Lv, Z, Lin, D, Zhu, B, Miao, Q, Bao, X, Shang, H. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of human USP7 USP domain from Biortus.
To Be Published
|
|
7CVP
 
 | | The Crystal Structure of human PHGDH from Biortus. | | Descriptor: | D-3-phosphoglycerate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Miao, Q, Huang, Y. | | Deposit date: | 2020-08-26 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of human PHGDH from Biortus.
To Be Published
|
|
7C4I
 
 | |
6WQ2
 
 | | Cryo-EM of the S. islandicus filamentous virus, SIFV | | Descriptor: | A-DNA, Structural protein MCP1, Structural protein MCP2 | | Authors: | Wang, F, Baquero, D.P, Su, Z, Zheng, W, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WQ0
 
 | | Cryo-EM of the S. solfataricus rod-shaped virus, SSRV1 | | Descriptor: | DNA (301-MER), Structural protein | | Authors: | Wang, F, Baquero, D.P, Beltran, L.C, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7D4A
 
 | | The Crystal Structure of human JMJD2A Tudor domain from Biortus | | Descriptor: | Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Ju, C, Bao, X, Zhu, B. | | Deposit date: | 2020-09-23 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | The Crystal Structure of human JMJD2A from Biortus.
To Be Published
|
|
7CMR
 
 | | The Crystal Structure of human MYST1 from Biortus. | | Descriptor: | GLYCEROL, Histone acetyltransferase KAT8, ZINC ION | | Authors: | Wang, F, Lin, D, Lv, Z, Xu, X, Tan, J, Shang, H. | | Deposit date: | 2020-07-28 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of human MYST1 from Biortus.
To Be Published
|
|
7C62
 
 | |
7CML
 
 | | The Crystal Structure of human JNK2 from Biortus. | | Descriptor: | Mitogen-activated protein kinase 9 | | Authors: | Wang, F, Lin, D, Cheng, W, Miao, Q, Huang, Y, Shang, H. | | Deposit date: | 2020-07-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of human JNK2 from Biortus.
To Be Published
|
|
7ESE
 
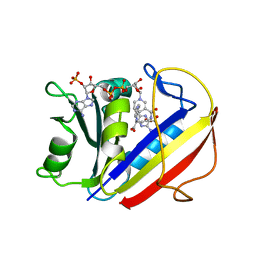 | | The Crystal Structure of human DHFR from Biortus | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, F, Cheng, W, Xu, C, Qi, J, Bao, X, Miao, Q. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of human DHFR from Biortus
To Be Published
|
|
7CA4
 
 | |
2Z2G
 
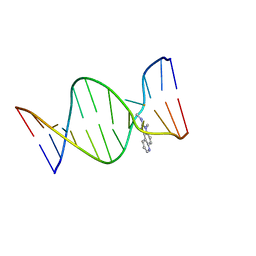 | | NMR Structure of the IQ-modified Dodecamer CTC[IQ]GGCGCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, DNA (5'-D(*DCP*DTP*DCP*DGP*DGP*DCP*DGP*DCP*DCP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DGP*DGP*DCP*DGP*DCP*DCP*DGP*DAP*DG)-3') | | Authors: | Wang, F, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme
Biochemistry, 46, 2007
|
|
2Z2H
 
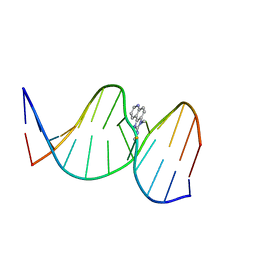 | | NMR Structure of the IQ-modified Dodecamer CTCG[IQ]GCGCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, DNA (5'-D(*DCP*DTP*DCP*DGP*DGP*DCP*DGP*DCP*DCP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DGP*DGP*DCP*DGP*DCP*DCP*DGP*DAP*DG)-3') | | Authors: | Wang, F, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme
Biochemistry, 46, 2007
|
|
2HKC
 
 | | NMR Structure of the IQ-modified Dodecamer CTCGGC[IQ]GCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, 5'-D(*CP*TP*CP*GP*GP*CP*GP*CP*CP*AP*TP*C)-3', 5'-D(*GP*AP*TP*GP*GP*CP*GP*CP*CP*GP*AP*G)-3' | | Authors: | Wang, F, DeMuro, N.E, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-03 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for -2 bp deletions.
J.Am.Chem.Soc., 128, 2006
|
|
2HKB
 
 | | NMR Structure of the B-DNA Dodecamer CTCGGCGCCATC | | Descriptor: | 5'-D(*CP*TP*CP*GP*GP*CP*GP*CP*CP*AP*TP*C)-3', 5'-D(*GP*AP*TP*GP*GP*CP*GP*CP*CP*GP*AP*G)-3' | | Authors: | Wang, F, DeMuro, N.E, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-03 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for -2 bp deletions.
J.Am.Chem.Soc., 128, 2006
|
|
2HVC
 
 | | The Crystal Structure of Ligand-binding Domain (LBD) of human Androgen Receptor in Complex with a selective modulator LGD2226 | | Descriptor: | 6-[BIS(2,2,2-TRIFLUOROETHYL)AMINO]-4-(TRIFLUOROMETHYL)QUINOLIN-2(1H)-ONE, Androgen receptor | | Authors: | Wang, F, Liu, X.-Q, Li, H, Liang, K.-N, Miner, J.N, Hong, M, Kallel, E.A, van Oeveren, A, Zhi, L, Jiang, T. | | Deposit date: | 2006-07-28 | | Release date: | 2007-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the ligand-binding domain (LBD) of human androgen receptor in complex with a selective modulator LGD2226
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
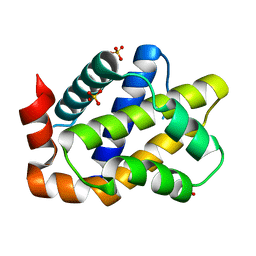
2K86
 
 | | Solution Structure of FOXO3a Forkhead domain | | Descriptor: | Forkhead box protein O3 | | Authors: | Wang, F, Marshall, C.B, Li, G, Plevin, M.J, Ikura, M. | | Deposit date: | 2008-09-02 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural characterization of an intramolecular interaction in FOXO3a and its binding with p53.
J.Mol.Biol., 384, 2008
|
|
