2QZU
 
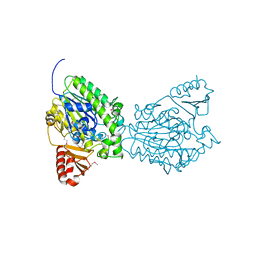 | | Crystal structure of the putative sulfatase yidJ from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR123 | | Descriptor: | Putative sulfatase yidJ | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Wang, D, Cunningham, K, Maglaqui, M, Owens, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-04 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystal structure of the putative sulfatase yidJ from Bacteroides fragilis.
To be Published
|
|
2P3Y
 
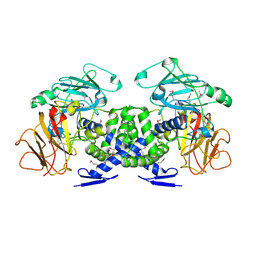 | | Crystal structure of VPA0735 from Vibrio parahaemolyticus. NorthEast Structural Genomics target VpR109 | | Descriptor: | Hypothetical protein VPA0735 | | Authors: | Seetharaman, J, Neely, H, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-10 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of VPA0735 from Vibrio parahaemolyticus.
To be Published
|
|
2RA8
 
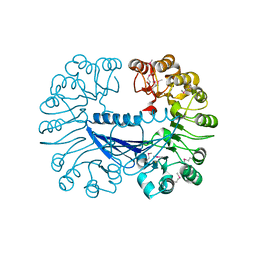 | | Crystal structure of the Q64V53_BACFR protein from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR43 | | Descriptor: | Uncharacterized protein Q64V53_BACFR | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Wang, D, Cunningham, K, Maglaqui, M, Owens, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the Q64V53_BACFR protein from Bacteroides fragilis.
To be Published
|
|
4MO1
 
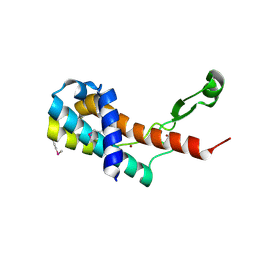 | | Crystal structure of antitermination protein Q from bacteriophage lambda. Northeast Structural Genomics Consortium target OR18A. | | Descriptor: | Antitermination protein Q, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Vorobiev, S, Su, M, Nickels, B, Seetharaman, J, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Wang, D, Everett, J.K, Acton, T.B, Ebright, R.H, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structure of antitermination protein Q from bacteriophage lambda.
To be Published
|
|
4N9X
 
 | | Crystal Structure of the OCTAPRENYL-METHYL-METHOXY-BENZQ MOLECULE from Erwina carotovora subsp. atroseptica strain SCRI 1043 / ATCC BAA-672, Northeast Structural Genomics Consortium (NESG) Target EwR161 | | Descriptor: | Putative monooxygenase | | Authors: | Kuzin, A, Chen, Y, Lew, S, Seetharaman, J, Mao, L, Xiao, R, Owens, L.A, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-21 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal Structure of the OCTAPRENYL-METHYL-METHOXY-BENZQ MOLECULE from Erwina carotovora subsp. atroseptica strain SCRI 1043 / ATCC BAA-672, Northeast Structural Genomics Consortium (NESG) Target EwR161
To be Published
|
|
4INA
 
 | | Crystal Structure of the Q7MSS8_WOLSU protein from Wolinella succinogenes. Northeast Structural Genomics Consortium Target WsR35 | | Descriptor: | saccharopine dehydrogenase | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Crystal Structure of the Q7MSS8_WOLSU protein from Wolinella succinogenes.
To be Published
|
|
3QSZ
 
 | | Crystal Structure of the STAR-related lipid transfer protein (fragment 25-204) from Xanthomonas axonopodis at the resolution 2.4A, Northeast Structural Genomics Consortium Target XaR342 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, STAR-related lipid transfer protein, ... | | Authors: | Kuzin, A.P, Su, M, Vorobiev, S.M, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | Northeast Structural Genomics Consortium Target XaR342
To be Published
|
|
4IOH
 
 | | Crystal Structure of the Tll1086 protein from Thermosynechococcus elongatus, Northeast Structural Genomics Consortium Target TeR258 | | Descriptor: | Tll1086 protein | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Maglaqui, M, Xiao, X, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-01-07 | | Release date: | 2013-01-30 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal Structure of the Tll1086 protein from Thermosynechococcus elongatus.
To be Published
|
|
2JPU
 
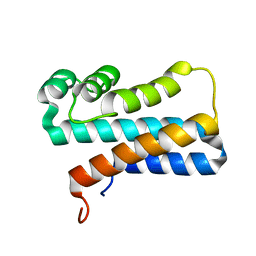 | | solution structure of NESG target SsR10, Orf c02003 protein | | Descriptor: | Orf c02003 protein | | Authors: | Wu, Y, Singarapu, K.K, Zhang, Q, Eletski, A, Xu, D, Sukumaran, D, Parish, D, Wang, D, Jiang, M, Cunningham, K, Maglaqui, M, Owens, L, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T, Rost, B, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-23 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of NESG target SsR10, Orf c02003 protein
To be Published
|
|
4TWK
 
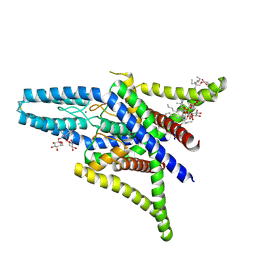 | | Crystal structure of human two pore domain potassium ion channel TREK1 (K2P2.1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Octyl Glucose Neopentyl Glycol, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Tessitore, A, Goubin, S, Strain-Damerell, C, Mukhopadhyay, S, Kupinska, K, Wang, D, Chalk, R, Berridge, G, Grieben, M, Shrestha, L, Ang, J.H, Mackenzie, A, Quigley, A, Bushell, S.R, Shintre, C.A, Faust, B, Chu, A, Dong, L, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Carpenter, E.P. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human two pore domain potassium ion channel TREK1 (K2P2.1)
To Be Published
|
|
3Q6C
 
 | | X-ray crystal structure of duf2500 (pf10694) from klebsiella pneumoniae, northeast structural genomics consortium target kpr96 | | Descriptor: | probable receptor YhhM | | Authors: | Seetharaman, J, Su, M, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-31 | | Release date: | 2011-08-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystal structure of duf2500 (pf10694) from klebsiella pneumoniae, northeast structural genomics consortium target kpr96
To be Published
|
|
3Q63
 
 | | X-ray crystal structure of protein MLL2253 from Mesorhizobium loti, Northeast Structural Genomics Consortium Target MlR404. | | Descriptor: | Mll2253 protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-30 | | Release date: | 2011-01-12 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of protein MLL2253 from Mesorhizobium loti, Northeast Structural Genomics Consortium Target MlR404.
To be Published
|
|
8I3X
 
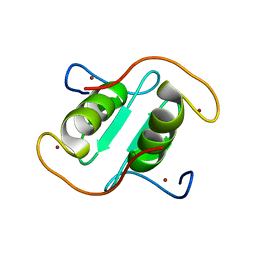 | | Rice APIP6-RING homodimer | | Descriptor: | RING-type domain-containing protein, ZINC ION | | Authors: | Zheng, Y, Zhang, X, Liu, Y, Liu, J, Wang, D. | | Deposit date: | 2023-01-18 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of rice APIP6 reveals a new dimerization mode of RING-type E3 ligases that facilities the construction of its working model
Phytopathol Res, 5, 2023
|
|
3RD6
 
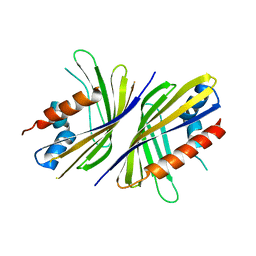 | | Crystal structure of Mll3558 protein from Rhizobium loti. Northeast Structural Genomics Consortium target id MlR403 | | Descriptor: | Mll3558 protein | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Ciccosanti, C, Sahdev, S, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-01 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Mll3558 protein from Rhizobium loti. Northeast Structural Genomics Consortium target id MlR403
To be Published
|
|
5YPI
 
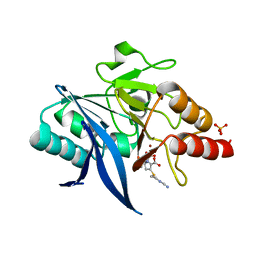 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EI1 complex | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPK
 
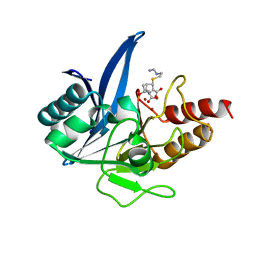 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EI2 complex | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPN
 
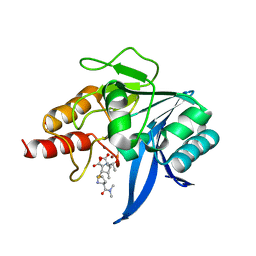 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI2 complex | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Liu, W, Wang, D. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPL
 
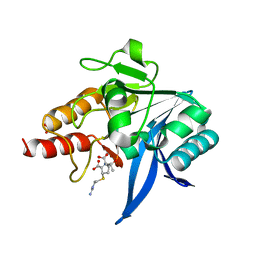 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EP complex | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPM
 
 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI1 complex | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
3SWV
 
 | | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A | | Descriptor: | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 | | Authors: | Seetharaman, J, Su, M, Forouhar, F, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-14 | | Release date: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BrefeldinA-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A
To be Published
|
|
3Q64
 
 | | X-ray crystal structure of protein mll3774 from Mesorhizobium loti, Northeast Structural Genomics Consortium Target MlR405. | | Descriptor: | Mll3774 protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-30 | | Release date: | 2011-01-12 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of protein mll3774 from Mesorhizobium loti, Northeast Structural Genomics Consortium Target MlR405.
To be Published
|
|
3Q6A
 
 | | X-ray crystal structure of the protein SSP2350 from Staphylococcus saprophyticus, Northeast structural genomics consortium target SyR116 | | Descriptor: | uncharacterized protein | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-31 | | Release date: | 2011-04-06 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of the protein SSP2350 from Staphylococcus saprophyticus, Northeast structural genomics consortium target SyR116
To be Published
|
|
3RD4
 
 | | Crystal structure of PROPEN_03304 from Proteus penneri ATCC 35198 Northeast Structural Genomics Consortium target id PvR55 | | Descriptor: | uncharacterized protein | | Authors: | Seetharaman, J, Min, S, Kuzin, A, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of PROPEN_03304 from Proteus penneri ATCC 35198 Northeast Structural Genomics Consortium target id PvR55
To be Published
|
|
8XGL
 
 | | Structure of the Magnaporthe oryzae effector NIS1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Necrosis-inducing secreted protein 1, ... | | Authors: | Han, R, Wang, D, Zhang, X, Liu, J. | | Deposit date: | 2023-12-15 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Understanding and manipulating the recognition of necrosis-inducing secreted protein 1 (NIS1) by BRI1-associated receptor kinase 1 (BAK1).
Int.J.Biol.Macromol., 278, 2024
|
|
6KMJ
 
 | | Crystal structure of Sth1 bromodomain in complex with H3K14Ac | | Descriptor: | GLYCEROL, Histone H3, Nuclear protein STH1/NPS1 | | Authors: | Chen, G, Li, W, Yan, F, Wang, D, Chen, Y. | | Deposit date: | 2019-07-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Structural Basis for Specific Recognition of H3K14 Acetylation by Sth1 in the RSC Chromatin Remodeling Complex.
Structure, 28, 2020
|
|
