7VNE
 
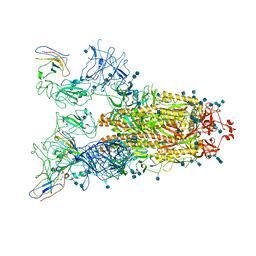 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113.1 (UUU-state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VNC
 
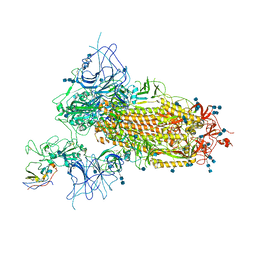 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UDD-state, state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VNB
 
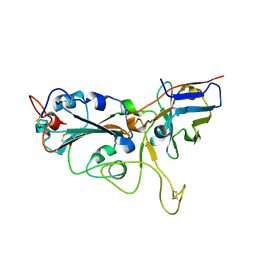 | | Crystal structure of the SARS-CoV-2 RBD in complex with a human single domain antibody n3113 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, n3113 | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VND
 
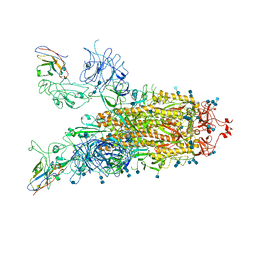 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UUD-state, state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
6LSN
 
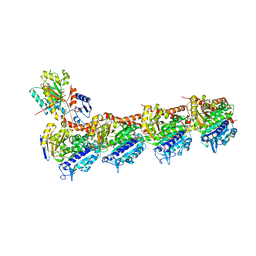 | | Crystal structure of tubulin-inhibitor complex | | Descriptor: | 2-(1-methylindol-5-yl)-7-(3,4,5-trimethoxyphenyl)pyrazolo[1,5-a]pyrimidine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Gang, L, Wang, Y.X, Cheng, J.J. | | Deposit date: | 2020-01-17 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Design, Synthesis, and Bioevaluation of Pyrazolo[1,5-a]Pyrimidine Derivatives as Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site with Potent Anticancer Activities
To Be Published
|
|
6VWV
 
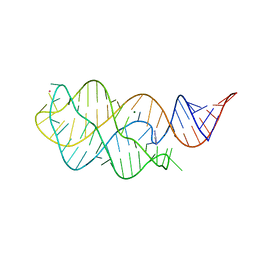 | |
6BRY
 
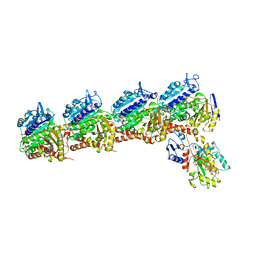 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 6a | | Descriptor: | 1-(2-chlorofuro[3,2-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
4N41
 
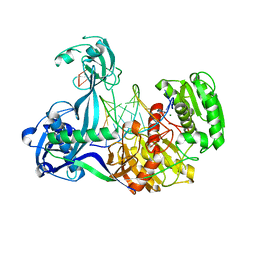 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 15-mer target DNA | | Descriptor: | 5'-D(*AP*AP*CP*CP*TP*AP*CP*TP*GP*CP*CP*TP*CP*G)-3', 5'-D(P*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*T*GP*TP*AP*TP*AP*GP*T)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6IIM
 
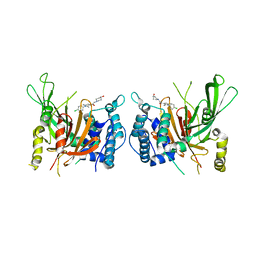 | | USP14 catalytic domain with IU1-206 | | Descriptor: | 1-[1-(4-chlorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(4-hydroxypiperidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, J.W, Wang, F, Wang, Y.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
6JV3
 
 | |
1BSI
 
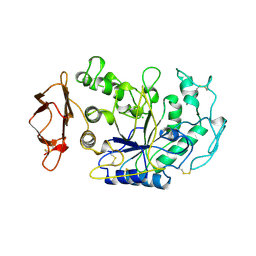 | | HUMAN PANCREATIC ALPHA-AMYLASE FROM PICHIA PASTORIS, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Rydberg, E.H, Sidhu, G, Vo, H.C, Hewitt, J, Cote, H.C.F, Wang, Y, Numao, S, Macgillivray, R.T.A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 1998-08-28 | | Release date: | 1999-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cloning, mutagenesis, and structural analysis of human pancreatic alpha-amylase expressed in Pichia pastoris.
Protein Sci., 8, 1999
|
|
3PR2
 
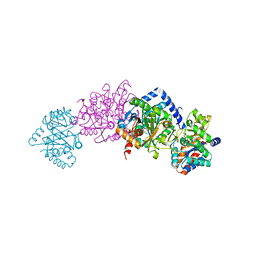 | | Tryptophan synthase indoline quinonoid structure with F9 inhibitor in alpha site | | Descriptor: | (Z)-N-[(1E)-1-carboxy-2-(2,3-dihydro-1H-indol-1-yl)ethylidene]{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methanaminium, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, CESIUM ION, ... | | Authors: | Lai, J, Niks, D, Wang, Y, Domratcheva, T, Barends, T.R.M, Schwarz, F, Olsen, R.A, Elliott, D.W, Fatmi, M.Q, Chang, C.A, Schlichting, I, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and NMR Crystallography in an Enzyme Active Site: The Indoline Quinonoid Intermediate in Tryptophan Synthase.
J.Am.Chem.Soc., 133, 2011
|
|
1ON3
 
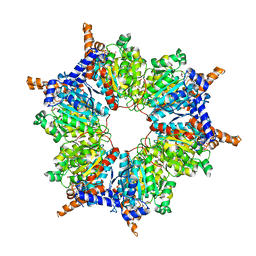 | | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with methylmalonyl-coenzyme a and methylmalonic acid bound) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CADMIUM ION, METHYLMALONIC ACID, ... | | Authors: | Hall, P.R, Wang, Y.-F, Rivera-Hainaj, R.E, Zheng, X, Pustai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core
Embo J., 22, 2003
|
|
4N76
 
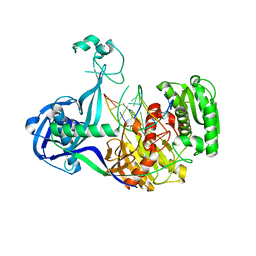 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and cleaved target DNA with Mn2+ | | Descriptor: | 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', Argonaute, ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-15 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1JNP
 
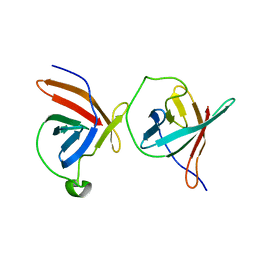 | | Crystal Structure of Murine Tcl1 at 2.5 Resolution | | Descriptor: | T-CELL LEUKEMIA/LYMPHOMA PROTEIN 1A | | Authors: | Petock, J.M, Torshin, I.Y, Wang, Y.F, DuBois, G.C, Croce, C.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2001-07-24 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of murine Tcl1 at 2.5 A resolution and implications for the TCL oncogene family.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6EEM
 
 | | Crystal structure of Papaver somniferum tyrosine decarboxylase in complex with L-tyrosine | | Descriptor: | N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-tyrosine, SULFATE ION, TYROSINE, ... | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.61000657 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEQ
 
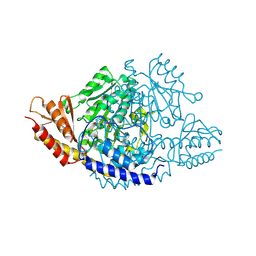 | | Crystal structure of Rhodiola rosea 4-hydroxyphenylacetaldehyde synthase | | Descriptor: | 4-hydroxyphenylacetaldehyde synthase | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.600086 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LY6
 
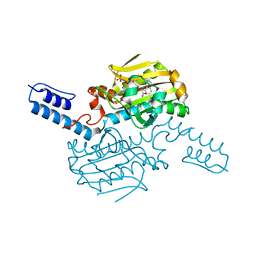 | | PylRS C-terminus domain mutant bound with 3-(1-Naphthyl)-L-alanine and AMPNP | | Descriptor: | MAGNESIUM ION, NAPHTHALEN-2-YL-3-ALANINE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5001328 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
6LY3
 
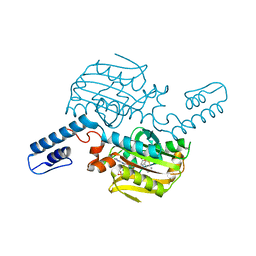 | | PylRS C-terminus domain mutant bound with 3-Benzothienyl-L-alanine and AMPNP | | Descriptor: | 3-(1-benzothiophen-3-yl)-L-alanine, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89590144 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
5YDG
 
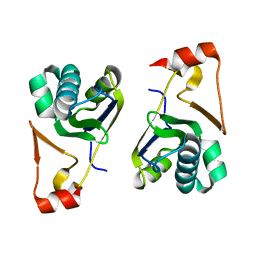 | | Crystal structure of the Arabidopsis thaliana chloroplast RNA editing factors 2(MORF2) | | Descriptor: | Multiple organellar RNA editing factor 2, chloroplastic | | Authors: | Wang, X, Yang, J.Y, Wang, Y.L, Gao, Y.S. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal structure of the chloroplast RNA editing factor MORF2
Biochem. Biophys. Res. Commun., 495, 2018
|
|
6LYA
 
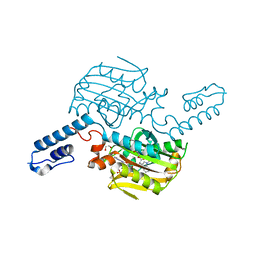 | | PylRS C-terminus domain mutant bound with 1-Methyl-L-tryptophan and AMPNP | | Descriptor: | 1,2-ETHANEDIOL, 1-Methyl-L-tryptophan, MAGNESIUM ION, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.590702 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
8W6K
 
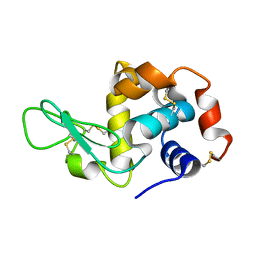 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2023-08-29 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | BL03HB: a Laue microdiffraction beamline for both protein crystallography and materials science at SSRF
Nucl.Sci.Tech., 35, 2024
|
|
2AOC
 
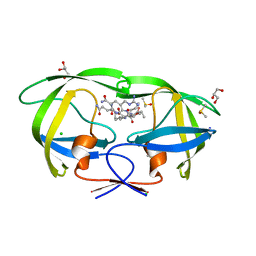 | | Crystal structure analysis of HIV-1 protease mutant I84V with a substrate analog P2-NC | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOG
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog P2-NC | | Descriptor: | ACETIC ACID, GLYCEROL, HIV-1 PROTEASE (RETROPEPSIN), ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
6LY7
 
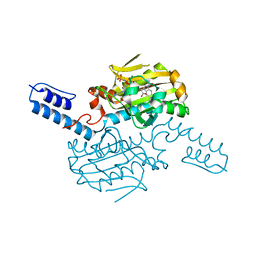 | | PylRS C-terminus domain mutant bound with 1-Formyl-L-tryptophan and AMPNP | | Descriptor: | MAGNESIUM ION, N1-FORMYL-TRYPTOPHAN, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09447265 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
