1K2C
 
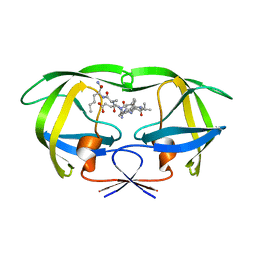 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | Deposit date: | 2001-09-26 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1K1U
 
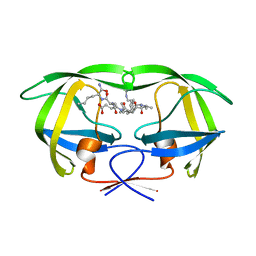 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1JR6
 
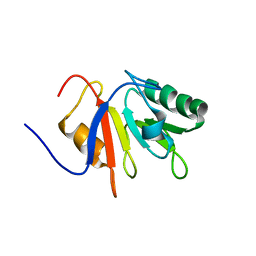 | |
1K2B
 
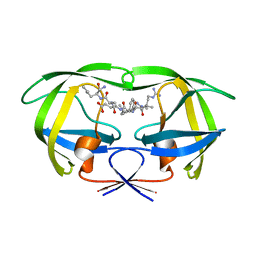 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | Deposit date: | 2001-09-26 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
5XWP
 
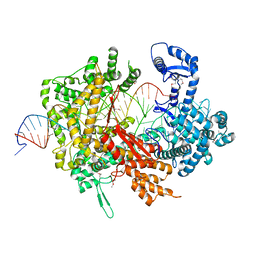 | | Crystal structure of LbuCas13a-crRNA-target RNA ternary complex | | Descriptor: | RNA (30-MER), RNA (59-MER), Uncharacterized protein | | Authors: | Liu, L, Li, X, Li, Z, Wang, Y. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
6WWC
 
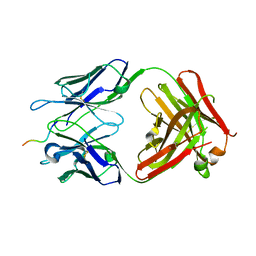 | |
6WX2
 
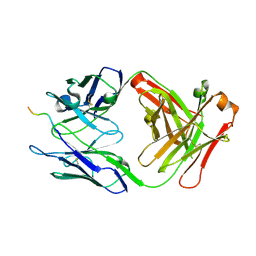 | |
6EG5
 
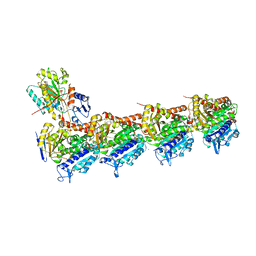 | | The structure of SB-1-202-tubulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chloropyrido[2,3-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | Banerjee, A.K, Wang, Y, Chen, H, Miller, D, Li, W. | | Deposit date: | 2018-08-18 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of SB-1-202/SB-2-204-tubulin complex
To Be Published
|
|
1TQH
 
 | | Covalent Reaction intermediate Revealed in Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est30 | | Descriptor: | Carboxylesterase precursor, PROPYL ACETATE, SULFATE ION | | Authors: | Liu, P, Wang, Y.F, Ewis, H.E, Abdelal, A.T, Lu, C.D, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Covalent reaction intermediate revealed in crystal structure of the Geobacillus stearothermophilus carboxylesterase Est30.
J.Mol.Biol., 342, 2004
|
|
7SPR
 
 | |
5YEL
 
 | | Crystal structure of CTCF ZFs6-11-gb7CSE | | Descriptor: | DNA (26-MER), Transcriptional repressor CTCF, ZINC ION | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-18 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
8EG5
 
 | | huCaspase-6 in complex with inhibitor 3a | | Descriptor: | (3R,5S)-1-(ethanesulfonyl)-5-phenyl-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide (bound form), 1,2-ETHANEDIOL, Caspase-6 subunit p11, ... | | Authors: | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | Deposit date: | 2022-09-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
1M2I
 
 | | Crystal structure of E44A/E56A mutant of cytochrome b5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Wu, J, Wang, Y.-H, Gan, J.-H, Wang, W.-H, Sun, B.-Y, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2002-06-24 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Cytochrome b5 Mutated at the Charged Surface-Residues and Their Interactions with Cytochrome c
Chin.J.Chem., 20, 2002
|
|
1M59
 
 | | Crystal Structure of P40V Mutant of Trypsin-solubilized Fragment of Cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Z.-Q, Wu, J, Wang, Y.-H, Qian, W, Xie, Y, Xia, Z.-X, Huang, Z.-X. | | Deposit date: | 2002-07-09 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proline40 is essential to maintaining cytochrome b5 stability and its electron transfer with cytochrome c
Chin.J.Chem., 20, 2002
|
|
5YEH
 
 | | Crystal structure of CTCF ZFs4-8-eCBS | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*CP*CP*GP*CP*TP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*AP*GP*CP*GP*GP*AP*AP*AP*CP*CP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
1YU7
 
 | | Crystal Structure of the W64Y mutant of Villin Headpiece | | Descriptor: | Villin | | Authors: | Meng, J, Vardar, D, Wang, Y, Guo, H.C, Head, J.F, McKnight, C.J. | | Deposit date: | 2005-02-12 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of villin headpiece and mutants with reduced F-actin binding activity.
Biochemistry, 44, 2005
|
|
1WMS
 
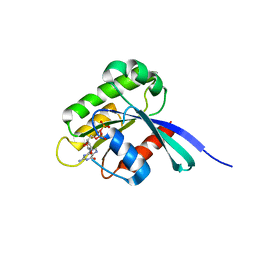 | | High resolution crystal structure of human Rab9 GTPase: a novel antiviral drug target | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-9A | | Authors: | Chen, L, DiGiammarino, E, Zhou, X.E, Wang, Y, Toh, D, Hodge, T.W, Meehan, E.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High resolution crystal structure of human Rab9 GTPase: A novel antiviral drug target
J.Biol.Chem., 279, 2004
|
|
6D88
 
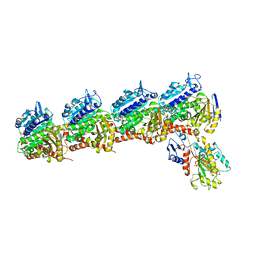 | | Tubulin-RB3_SLD-TTL in complex with compound 13f | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2018-04-26 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural Modification of the 3,4,5-Trimethoxyphenyl Moiety in the Tubulin Inhibitor VERU-111 Leads to Improved Antiproliferative Activities.
J. Med. Chem., 61, 2018
|
|
1KGU
 
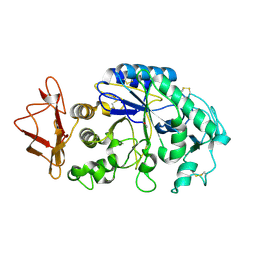 | | THREE DIMENSIONAL STRUCTURE ANALYSIS OF THE R337A VARIANT OF HUMAN PANCREATIC ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CALCIUM ION | | Authors: | Numao, S, Maurus, R, Sidhu, G, Wang, Y, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-28 | | Release date: | 2002-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of the chloride ion in the mechanism of human pancreatic alpha-amylase.
Biochemistry, 41, 2002
|
|
1X6L
 
 | | Crystal structure of S. marcescens chitinase A mutant W167A | | Descriptor: | Chitinase A | | Authors: | Aronson Jr, N.N, Halloran, B.A, Alexyev, M.F, Zhou, X.E, Wang, Y, Meehan, E.J, Chen, L. | | Deposit date: | 2004-08-11 | | Release date: | 2005-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutation of Trp167 at the -3 subsite of the chitin-binding cleft of S. marcescens chitinase A caused enhanced transglycosylation
To be Published
|
|
1X6N
 
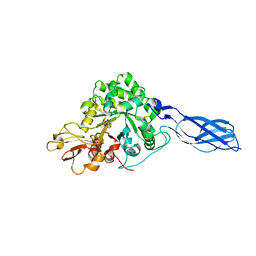 | | Crystal structure of S. marcescens chitinase A mutant W167A in complex with allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, Chitinase A | | Authors: | Aronson Jr, N.N, Halloran, B.A, Alexyev, M.F, Zhou, X.E, Wang, Y, Meehan, E.J, Chen, L. | | Deposit date: | 2004-08-11 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Muation of Trp167 at the -3 subsite of the chitin-binding cleft of S. marcescens chitinase A causes enhanced transglycosylation
To be Published
|
|
1ZUB
 
 | | Solution Structure of the RIM1alpha PDZ Domain in Complex with an ELKS1b C-terminal Peptide | | Descriptor: | ELKS1b, Regulating synaptic membrane exocytosis protein 1 | | Authors: | Lu, J, Li, H, Wang, Y, Sudhof, T.C, Rizo, J. | | Deposit date: | 2005-05-30 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RIM1alpha PDZ Domain in Complex with an ELKS1b C-terminal Peptide
J.Mol.Biol., 352, 2005
|
|
1NA6
 
 | | Crystal structure of restriction endonuclease EcoRII mutant R88A | | Descriptor: | Restriction endonuclease EcoRII | | Authors: | Zhou, X.E, Wang, Y, Reuter, M, Mucke, M, Kruger, D.H, Meehan, E.J, Chen, L. | | Deposit date: | 2002-11-26 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold.
J.Mol.Biol., 335, 2004
|
|
6IFO
 
 | | Crystal structure of AcrIIA2-SpyCas9-sgRNA ternary complex | | Descriptor: | AcrIIA2, CRISPR-associated endonuclease Cas9/Csn1, RNA (99-MER) | | Authors: | Liu, L, Wang, Y. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Phage AcrIIA2 DNA Mimicry: Structural Basis of the CRISPR and Anti-CRISPR Arms Race.
Mol. Cell, 73, 2019
|
|
8K3G
 
 | |
