4FJ4
 
 | | Crystal structure of the protein Q9HRE7 complexed with mercury from Halobacterium salinarium at the resolution 2.1A, Northeast Structural Genomics Consortium target HsR50 | | Descriptor: | ETHYL MERCURY ION, SODIUM ION, Uncharacterized protein | | Authors: | Kuzin, A, Chen, Y, Vorobiev, S.M, Seetharaman, J, Janjua, J, Xiao, R, Cunningham, K, Maglaqui, M, Owens, L.A, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Northeast Structural Genomics Consortium Target HsR50
To be Published
|
|
1Q2I
 
 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | Descriptor: | PNC27 | | Authors: | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | Deposit date: | 2003-07-24 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
3NGW
 
 | | Crystal Structure of Molybdopterin-guanine dinucleotide biosynthesis protein A from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium Target GR189 | | Descriptor: | CALCIUM ION, Molybdopterin-guanine dinucleotide biosynthesis protein A (MobA) | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-13 | | Release date: | 2010-08-11 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Northeast Structural Genomics Consortium Target GR189
To be Published
|
|
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4YGQ
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | Hydrolase, TERTIARY-BUTYL ALCOHOL | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
7D0B
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-3C12_1B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3C12, ... | | Authors: | Yan, R.H, Wang, R.K, Ju, B, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
4YGR
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
5WH6
 
 | | Crystal structure of PDE4D2 in complex with inhibitor (S_Zl-n-91) | | Descriptor: | 1-[4-(difluoromethoxy)-3-{[(3S)-oxolan-3-yl]oxy}phenyl]-3-methylbutan-1-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ke, H, Wang, H. | | Deposit date: | 2017-07-14 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a PDE4-Specific Pocket for the Design of Selective Inhibitors.
Biochemistry, 57, 2018
|
|
2KCT
 
 | | Solution nmr structure of the ob-fold domain of heme chaperone ccme from desulfovibrio vulgaris. northeast structural genomics target dvr115g. | | Descriptor: | Cytochrome c-type biogenesis protein CcmE | | Authors: | Aramini, J.M, Rossi, P, Lee, H, Lemak, A, Wang, H, Foote, E.L, Jiang, M, Xiao, R, Nair, R, Swapna, G.V.T, Acton, T.B, Rost, B, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution nmr structure of the ob-fold domain of heme chaperone
ccme from desulfovibrio vulgaris. northeast structural genomics target dvr115g.
To be Published
|
|
1Q2F
 
 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | Descriptor: | PNC27 | | Authors: | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | Deposit date: | 2003-07-24 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
4YGS
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | CITRIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
3G3N
 
 | | PDE7A catalytic domain in complex with 3-(2,6-difluorophenyl)-2-(methylthio)quinazolin-4(3H)-one | | Descriptor: | 3-(2,6-difluorophenyl)-2-(methylthio)quinazolin-4(3H)-one, High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | Authors: | Castano, T, Wang, H. | | Deposit date: | 2009-02-02 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis, structural analysis, and biological evaluation of thioxoquinazoline derivatives as phosphodiesterase 7 inhibitors
Chemmedchem, 4, 2009
|
|
5Z1Q
 
 | | Crystal structures of the trimeric N-terminal domain of Ciliate Euplotes octocarinatus Centrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Centrin protein | | Authors: | Wang, W, Zhao, Y, Yang, B, Wang, H. | | Deposit date: | 2017-12-27 | | Release date: | 2018-04-25 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the trimeric N-terminal domain of ciliate Euplotes octocarinatus centrin binding with calcium ions
Protein Sci., 27, 2018
|
|
2KO1
 
 | | Solution NMR structure of the ACT domain from GTP pyrophosphokinase of Chlorobium tepidum. Northeast Structural Genomics Consortium Target CtR148A | | Descriptor: | GTP pyrophosphokinase | | Authors: | Eletsky, A, Garcia, E, Wang, H, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the ACT domain from GTP pyrophosphokinase of Chlorobium tepidum
To be Published
|
|
2CWE
 
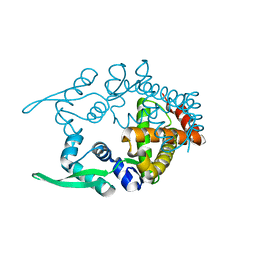 | | Crystal structure of hypothetical transcriptional regulator protein, PH1932 from Pyrococcus horikoshii OT3 | | Descriptor: | hypothetical transcription regulator protein, PH1932 | | Authors: | Arai, R, Kishishita, S, Kukimoto-Niino, M, Wang, H, Sugawara, M, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-20 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical transcriptional regulator protein, PH1932 from Pyrococcus horikoshii OT3
To be Published
|
|
6K41
 
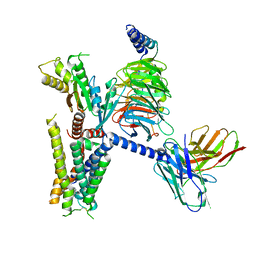 | | cryo-EM structure of alpha2BAR-GoA complex | | Descriptor: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor,Endolysin,Alpha-2B adrenergic receptor,Alpha-2B adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, D, Liu, Z, Wang, H.W, Kobilka, B.K. | | Deposit date: | 2019-05-23 | | Release date: | 2020-04-15 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of the alpha2Badrenoceptor by the sedative sympatholytic dexmedetomidine.
Nat.Chem.Biol., 16, 2020
|
|
6K42
 
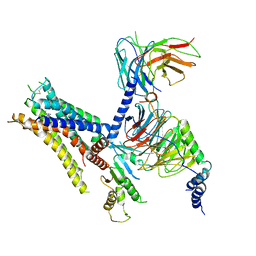 | | cryo-EM structure of alpha2BAR-Gi1 complex | | Descriptor: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor,Endolysin,Alpha-2B adrenergic receptor,Alpha-2B adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, D, Liu, Z, Wang, H.W, Kobilka, B.K. | | Deposit date: | 2019-05-23 | | Release date: | 2020-04-15 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Activation of the alpha2Badrenoceptor by the sedative sympatholytic dexmedetomidine.
Nat.Chem.Biol., 16, 2020
|
|
2ZM6
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kaminishi, T, Wang, H, Kawazoe, M, Ishii, R, Schluenzen, F, Hanawa-Suetsugu, K, Wilson, D.N, Nomura, M, Takemoto, C, Shirouzu, M, Fucini, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-11 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the Thermus thermophilus 30S ribosomal subunit
To be Published
|
|
7EVP
 
 | | Cryo-EM structure of the Gp168-beta-clamp complex | | Descriptor: | Beta sliding clamp, Sliding clamp inhibitor | | Authors: | Liu, B, Li, S, Liu, Y, Chen, H, Hu, Z, Wang, Z, Gou, L, Zhang, L, Ma, B, Wang, H, Matthews, S, Wang, Y, Zhang, K. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage Twort protein Gp168 is a beta-clamp inhibitor by occupying the DNA sliding channel.
Nucleic Acids Res., 49, 2021
|
|
7FH6
 
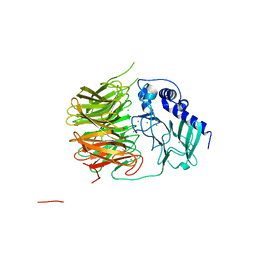 | | Friedel-Crafts alkylation enzyme CylK | | Descriptor: | CALCIUM ION, CHLORIDE ION, CylK, ... | | Authors: | Liu, L, Wang, H.Q, Xiang, Z. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for the Friedel-Crafts Alkylation in Cylindrocyclophane Biosynthesis
ACS Catal., 12, 2022
|
|
3TYH
 
 | | Crystal structure of oxo-cupper clusters binding to ferric binding protein from Neisseria gonorrhoeae | | Descriptor: | COPPER (II) ION, FbpA protein | | Authors: | Chen, W.J, Wang, H.F, Zhou, C.J, Ye, D.R, Huang, J, Tan, X.S, Zhong, W.Q. | | Deposit date: | 2011-09-26 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of oxo-cupper clusters binding to ferric binding protein from Neisseria gonorrhoeae
To be Published
|
|
4C5Y
 
 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE, ZINC ION | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
2LG3
 
 | | Structure of the duplex containing HNE derived (6S,8R,11S) gamma-HO-PdG when placed opposite dT | | Descriptor: | (4S)-nonane-1,4-diol, DNA (5'-D(*GP*CP*TP*AP*GP*CP*GP*AP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*GP*CP*TP*AP*GP*C)-3') | | Authors: | Huang, H, Wang, H, Kozekova, A, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ring-chain tautomerization of trans-4-hydroxynonenal derived (6S,8R,11S) gamma-hydroxy-1,N2-propano-deoxyguanosine adduct when placed opposite 2'-deoxythymidine in duplex
To be Published
|
|
8T8M
 
 | | Quis-bound intermediate mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nb43 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-22 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8T6J
 
 | | CDPPB-bound inactive mGlu5 | | Descriptor: | 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-16 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
