2PRL
 
 | | The structures of apo- and inhibitor bound human dihydroorotate dehydrogenase reveal conformational flexibility within the inhibitor binding site | | Descriptor: | 5-METHOXY-2-[(4-PHENOXYPHENYL)AMINO]BENZOIC ACID, ACETATE ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Walse, B, Dufe, V.T, Al-Karadaghi, S. | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structures of human dihydroorotate dehydrogenase with and without inhibitor reveal conformational flexibility in the inhibitor and substrate binding sites
Biochemistry, 47, 2008
|
|
2PRH
 
 | | The structures of apo- and inhibitor bound human dihydroorotate dehydrogenase reveal conformational flexibility within the inhibitor binding site | | Descriptor: | 6-CHLORO-2-(2'-FLUOROBIPHENYL-4-YL)-3-METHYLQUINOLINE-4-CARBOXYLIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, Dihydroorotate dehydrogenase, ... | | Authors: | Walse, B, Dufe, V.T, Al-Karadaghi, S. | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structures of human dihydroorotate dehydrogenase with and without inhibitor reveal conformational flexibility in the inhibitor and substrate binding sites
Biochemistry, 47, 2008
|
|
2WV8
 
 | | Complex of human dihydroorotate dehydrogenase with the inhibitor 221290 | | Descriptor: | 2-ACETAMIDO-5-(4-PHENYLPHENYL)BENZOIC ACID, ACETATE ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Walse, B, Svensson, B, Fritzson, I, Dahlberg, L, Wellmar, U, Al-Karadaghi, S. | | Deposit date: | 2009-10-15 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of Human Dhodh by 4-Hydroxycoumarins, Fenamic Acids, and N-(Alkylcarbonyl)Anthranilic Acids Identified by Structure-Guided Fragment Selection.
Chemmedchem, 5, 2010
|
|
2PRM
 
 | | The structures of apo- and inhibitor bound human dihydroorotate dehydrogenase reveal conformational flexibility within the inhibitor binding site | | Descriptor: | Dihydroorotate dehydrogenase, mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Walse, B, Dufe, V.T, Al-Karadaghi, S. | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of human dihydroorotate dehydrogenase with and without inhibitor reveal conformational flexibility in the inhibitor and substrate binding sites
Biochemistry, 47, 2008
|
|
1HXY
 
 | | CRYSTAL STRUCTURE OF STAPHYLOCOCCAL ENTEROTOXIN H IN COMPLEX WITH HUMAN MHC CLASS II | | Descriptor: | ENTEROTOXIN H, HEMAGGLUTININ, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Petersson, K, Hakansson, M, Nilsson, H, Forsberg, G, Svensson, L.A, Liljas, A, Walse, B. | | Deposit date: | 2001-01-17 | | Release date: | 2001-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Superantigen Bound to MHC Class II Displays Zinc and Peptide Dependence
Embo J., 20, 2001
|
|
8P4Z
 
 | | Crystal structure of the human CDK7 kinase domain in complex with LDC4297 | | Descriptor: | 2-[(3R)-piperidin-3-yl]oxy-8-propan-2-yl-N-[(2-pyrazol-1-ylphenyl)methyl]pyrazolo[1,5-a][1,3,5]triazin-4-amine, Cyclin-dependent kinase 7, GLYCEROL, ... | | Authors: | Laursen, M, Caing-Carlsson, R, Houssari, R, Javadi, A, Kimbung, Y.R, Murina, V, Orozco-Rodriguez, J.M, Svensson, A, Welin, M, Logan, D, Svensson, B, Walse, B. | | Deposit date: | 2023-05-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the human CDK7 kinase domain in complex with LDC4297
To Be Published
|
|
9EOU
 
 | |
5MPR
 
 | | Single Amino Acid Variant of Human Mitochondrial Branched Chain Amino Acid Aminotransferase 2 | | Descriptor: | 1,2-ETHANEDIOL, Branched-chain-amino-acid aminotransferase, mitochondrial, ... | | Authors: | Hakansson, M, Walse, B, Nilsson, C, Anderson, L.C. | | Deposit date: | 2016-12-18 | | Release date: | 2017-07-19 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Intact Protein Analysis at 21 Tesla and X-Ray Crystallography Define Structural Differences in Single Amino Acid Variants of Human Mitochondrial Branched-Chain Amino Acid Aminotransferase 2 (BCAT2).
J. Am. Soc. Mass Spectrom., 28, 2017
|
|
6YKD
 
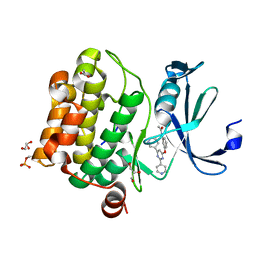 | | Human Pim-1 kinase in complex with an inhibitor identified by virtual screening | | Descriptor: | ACETATE ION, GLYCEROL, Serine/threonine-protein kinase pim-1, ... | | Authors: | Schneider, P, Welin, M, Svensson, B, Walse, B, Schneider, G. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Virtual Screening and Design with Machine Intelligence Applied to Pim-1 Kinase Inhibitors.
Mol Inform, 39, 2020
|
|
1LO5
 
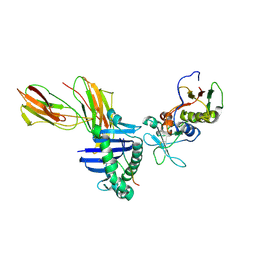 | | Crystal structure of the D227A variant of Staphylococcal enterotoxin A in complex with human MHC class II | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DR-1 beta chain, ... | | Authors: | Petersson, K, Thunnissen, M, Forsberg, G, Walse, B. | | Deposit date: | 2002-05-06 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of a SEA Variant in Complex with MHC Class II Reveals the Ability of SEA to Crosslink MHC Molecules
Structure, 10, 2002
|
|
7TPS
 
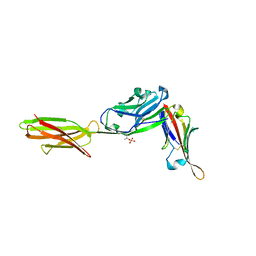 | | Crystal structure of ALPN-202 (engineered CD80 vIgD) in complex with PD-L1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Demonte, D.W, Maurer, M.F, Akutsu, M, Kimbung, Y.R, Logan, D.T, Walse, B. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The engineered CD80 variant fusion therapeutic davoceticept combines checkpoint antagonism with conditional CD28 costimulation for anti-tumor immunity.
Nat Commun, 13, 2022
|
|
6GK0
 
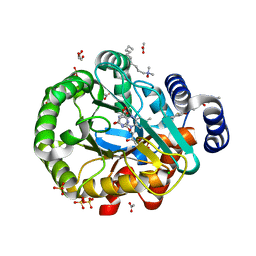 | | HUMAN DIHYDROOROTATE DEHYDROGENASE IN COMPLEX WITH CLASS III HISTONE DEACETYLASE INHIBITOR | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 4-~{tert}-butyl-~{N}-[[4-[5-(dimethylamino)pentanoylamino]phenyl]carbamothioyl]benzamide, ACETIC ACID, ... | | Authors: | Hakansson, M, Ladds, M.J.G.W, Walse, B, Lain, S. | | Deposit date: | 2018-05-17 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploitation of dihydroorotate dehydrogenase (DHODH) and p53 activation as therapeutic targets: A case study in polypharmacology.
J.Biol.Chem., 295, 2020
|
|
6ET4
 
 | | HUMAN DIHYDROOROTATE DEHYDROGENASE IN COMPLEX WITH NOVEL INHIBITOR | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Hakansson, M, Walse, B, Gustavsson, A.-L, Lain, S. | | Deposit date: | 2017-10-25 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A DHODH inhibitor increases p53 synthesis and enhances tumor cell killing by p53 degradation blockage.
Nat Commun, 9, 2018
|
|
5A6N
 
 | | Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with compound 2 | | Descriptor: | 5-(3-SULFAMOYLPHENYL)-1H-1,2,3,4-TETRAZOL-1-IDE, DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, ... | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6S4G
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruggieri, F, Campillo Brocal, J.C, Humble, M.S, Walse, B, Logan, D.T, Berglund, P. | | Deposit date: | 2019-06-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Insight into the dimer dissociation process of the Chromobacterium violaceum (S)-selective amine transaminase.
Sci Rep, 9, 2019
|
|
5A6O
 
 | | Crystal structure of the apo form of the unphosphorylated human death associated protein kinase 3 (DAPK3) | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, S-1,2-PROPANEDIOL | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6SNU
 
 | | Crystal structure of the W60C mutant of the (S)-selective transaminase from Chromobacterium violaceum | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase family protein, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ruggieri, F, Gustafsson, C, Kimbung, R.Y, Walse, B, Logan, D.T, Berglund, P. | | Deposit date: | 2019-08-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Combined with Molecular Dynamics Reveal Altered Flow of Water in the Active Site of W60C Chromobacterium violaceum omega-transaminase
Not Published
|
|
5A7C
 
 | | Crystal structure of the second bromodomain of human BRD3 in complex with compound | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 3, N-(6-ACETAMIDOHEXYL)ACETAMIDE | | Authors: | Welin, M, Kimbung, R, Diehl, C, Hakansson, M, Logan, D.T, Walse, B. | | Deposit date: | 2015-07-03 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cancer Differentiating Agent Hexamethylene Bisacetamide Inhibits Bet Bromodomain Proteins.
Cancer Res., 76, 2016
|
|
4A6R
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in the apo form, crystallised from polyacrylic acid | | Descriptor: | OMEGA TRANSAMINASE, POLYACRYLIC ACID | | Authors: | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | Deposit date: | 2011-11-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
4A6U
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in the apo form, crystallised from PEG 3350 | | Descriptor: | OMEGA TRANSAMINASE, SODIUM ION, THIOCYANATE ION | | Authors: | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | Deposit date: | 2011-11-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
4A6T
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in complex with PLP | | Descriptor: | OMEGA TRANSAMINASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | Deposit date: | 2011-11-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
4A72
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in a mixture of apo and PLP-bound states | | Descriptor: | OMEGA TRANSAMINASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | Deposit date: | 2011-11-10 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
2Y6H
 
 | | X-2 L110F CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
2Y6J
 
 | | X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
2Y6G
 
 | | Cellopentaose binding mutated (X-2 L110F) CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
