7E2E
 
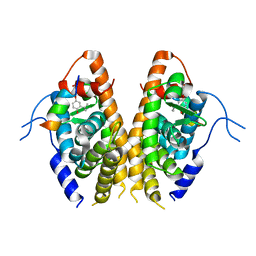 | | Crystal structure of the Estrogen-Related Receptor alpha (ERRalpha) ligand-binding domain (LBD) in complex with an agonist DS45500853 and a PGC-1alpha peptide | | Descriptor: | 1-[4-(3-tert-butyl-4-oxidanyl-phenoxy)phenyl]ethanone, IODIDE ION, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Ito, S, Shinozuka, T, Kimura, T, Izumi, M, Wakabayashi, K. | | Deposit date: | 2021-02-05 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of a Novel Class of ERR alpha Agonists.
Acs Med.Chem.Lett., 12, 2021
|
|
1IQA
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MOUSE RANK LIGAND | | Descriptor: | RECEPTOR ACTIVATOR OF NUCLEAR FACTOR KAPPA B LIGAND | | Authors: | Ito, S, Wakabayashi, K, Ubukata, O, Hayashi, S, Okada, F, Hata, T. | | Deposit date: | 2001-07-11 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the extracellular domain of mouse RANK ligand at 2.2-A resolution.
J.Biol.Chem., 277, 2002
|
|
5H6L
 
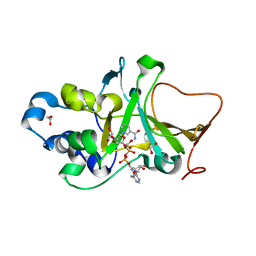 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6M
 
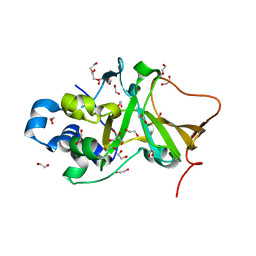 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6N
 
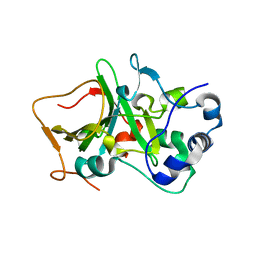 | | DNA targeting ADP-ribosyltransferase Pierisin-1, autoinhibitory form | | Descriptor: | Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6J
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6K
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
2EFS
 
 | | Crystal structure of the C-terminal tropomyosin fragment with N- and C-terminal extensions of the leucine zipper at 2.0 angstroms resolution | | Descriptor: | General control protein GCN4 and Tropomyosin 1 alpha chain | | Authors: | Minakata, S, Nitanai, Y, Maeda, K, Oda, N, Wakabayashi, K, Maeda, Y. | | Deposit date: | 2007-02-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two crystal structures of tropomyosin C-terminal fragment 176-273: exposure of the hydrophobic core to the solvent destabilizes the tropomyosin molecule
To be Published
|
|
2EFR
 
 | | Crystal structure of the c-terminal tropomyosin fragment with N- and C-terminal extensions of the leucine zipper at 1.8 angstroms resolution | | Descriptor: | General control protein GCN4 and Tropomyosin 1 alpha chain | | Authors: | Minakata, S, Nitanai, Y, Maeda, K, Oda, N, Wakabayashi, K, Maeda, Y. | | Deposit date: | 2007-02-23 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two crystal structures of tropomyosin C-terminal fragment 176-273: exposure of the hydrophobic core to the solvent destabilizes the tropomyosin molecule
To be Published
|
|
3B0W
 
 | | Crystal structure of the orphan nuclear receptor ROR(gamma)t ligand-binding domain in complex with digoxin | | Descriptor: | DIGOXIN, Nuclear receptor ROR-gamma | | Authors: | Fujita-Sato, S, Ito, S, Isobe, T, Ohyama, T, Wakabayashi, K, Morishita, K, Ando, O, Isono, F. | | Deposit date: | 2011-06-17 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Digoxin That Antagonizes ROR{gamma}t Receptor Activity and Suppresses Th17 Cell Differentiation and Interleukin (IL)-17 Production
J.Biol.Chem., 286, 2011
|
|
