3EP0
 
 | | Methyltransferase domain of human PR domain-containing protein 12 | | Descriptor: | PR domain zinc finger protein 12 | | Authors: | Amaya, M.F, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of methyltransferase domain of human PR
Domain-containing protein 12.
To be Published
|
|
4XCX
 
 | | METHYLTRANSFERASE DOMAIN OF SMALL RNA 2'-O-METHYLTRANSFERASE | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Small RNA 2'-O-methyltransferase | | Authors: | Walker, J.R, Zeng, H, Dong, A, Li, Y, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of human C1ORF59 in complex with SAH
To be published
|
|
3F2K
 
 | | Structure of the transposase domain of human Histone-lysine N-methyltransferase SETMAR | | Descriptor: | Histone-lysine N-methyltransferase SETMAR, LYFA Peptide, MAGNESIUM ION | | Authors: | Amaya, M.F, Dombrovski, L, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of Transposase Domain of Human
Histone-lysine N-methyltransferase SETMAR.
To be Published
|
|
4XQ1
 
 | | Crystal structure of hemerythrin: L114A mutant | | Descriptor: | Bacteriohemerythrin, FE (III) ION, NITRATE ION, ... | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
4XPW
 
 | | Crystal structures of Leu114F mutant | | Descriptor: | Bacteriohemerythrin, FE (II) ION, GLYCEROL | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
1ZQ9
 
 | | Crystal structure of human Dimethyladenosine transferase | | Descriptor: | Probable dimethyladenosine transferase, S-ADENOSYLMETHIONINE | | Authors: | Dong, A, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Plotnikov, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-18 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human Dimethyladenosine transferase with SAM
TO BE PUBLISHED
|
|
4XPY
 
 | | Crystal structure of hemerythrin : L114Y mutant | | Descriptor: | Bacteriohemerythrin, FE (II) ION, GLYCEROL | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
1XRK
 
 | | Crystal structure of a mutant bleomycin binding protein from Streptoalloteichus hindustanus displaying increased thermostability | | Descriptor: | BLEOMYCIN A2, Bleomycin resistance protein, SULFATE ION | | Authors: | Brouns, S.J.J, Wu, H, Akerboom, J, Turnbull, A.P, de Vos, W.M, Van der Oost, J. | | Deposit date: | 2004-10-15 | | Release date: | 2005-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering a selectable marker for hyperthermophiles
J.Biol.Chem., 280, 2005
|
|
3OQ9
 
 | | Structure of the FAS/FADD death domain assembly | | Descriptor: | Protein FADD, Tumor necrosis factor receptor superfamily member 6 | | Authors: | Kabaleeswaran, V, Wu, H. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6.8 Å) | | Cite: | The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3FLG
 
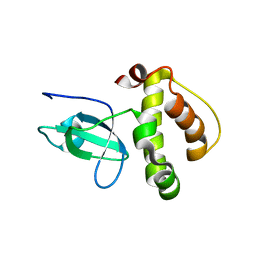 | | The PWWP domain of Human DNA (cytosine-5-)-methyltransferase 3 beta | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B | | Authors: | Amaya, M.F, Zeng, H, MacKenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the PWWP domain of Human DNA (cytosine-5-)-methyltransferase 3 beta
To be Published
|
|
6CBX
 
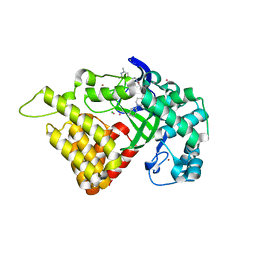 | | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF1497 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-lysine methyltransferase SMYD2, ... | | Authors: | ZENG, H, DONG, A, Hutchinson, A, Seitova, A, TATLOCK, J, KUMPF, R, OWEN, A, TAYLOR, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF1497
to be published
|
|
1WC3
 
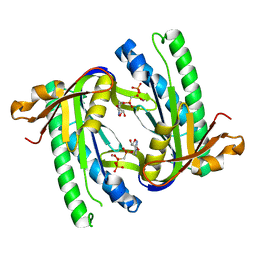 | | Soluble adenylyl cyclase CyaC from S. platensis in complex with alpha, beta-methylene-ATP and Strontium | | Descriptor: | ADENYLATE CYCLASE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, STRONTIUM ION | | Authors: | Steegborn, C, Litvin, T.N, Levin, L.R, Buck, J, Wu, H. | | Deposit date: | 2004-11-08 | | Release date: | 2004-12-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bicarbonate Activation of Adenylyl Cyclase Via Promotion of Catalytic Active Site Closure and Metal Recruitment
Nat.Struct.Mol.Biol., 12, 2005
|
|
1WC5
 
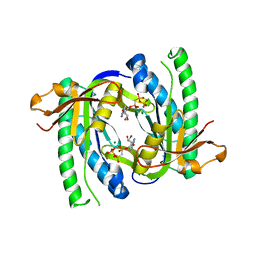 | | Soluble adenylyl cyclase CyaC from S. platensis in complex with alpha, beta-methylene-ATP in presence of bicarbonate | | Descriptor: | ADENYLATE CYCLASE, CALCIUM ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Steegborn, C, Litvin, T.N, Levin, L.R, Buck, J, Wu, H. | | Deposit date: | 2004-11-08 | | Release date: | 2004-12-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bicarbonate Activation of Adenylyl Cyclase Via Promotion of Catalytic Active Site Closure and Metal Recruitment
Nat.Struct.Mol.Biol., 12, 2005
|
|
1WC6
 
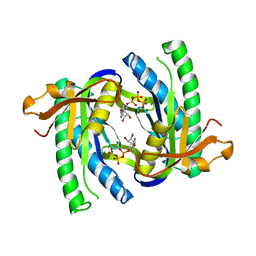 | | Soluble adenylyl cyclase CyaC from S. platensis in complex with Rp- ATPalphaS in presence of bicarbonate | | Descriptor: | ADENOSINE-5'-RP-ALPHA-THIO-TRIPHOSPHATE, ADENYLATE CYCLASE, MAGNESIUM ION | | Authors: | Steegborn, C, Litvin, T.N, Levin, L.R, Buck, J, Wu, H. | | Deposit date: | 2004-11-08 | | Release date: | 2004-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Bicarbonate Activation of Adenylyl Cyclase Via Promotion of Catalytic Active Site Closure and Metal Recruitment
Nat.Struct.Mol.Biol., 12, 2005
|
|
1WC1
 
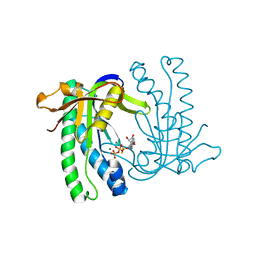 | | Soluble adenylyl cyclase CyaC from S. platensis in complex with Rp- ATPalphaS | | Descriptor: | ADENOSINE-5'-RP-ALPHA-THIO-TRIPHOSPHATE, ADENYLATE CYCLASE, MAGNESIUM ION | | Authors: | Steegborn, C, Litvin, T.N, Levin, L.R, Buck, J, Wu, H. | | Deposit date: | 2004-11-06 | | Release date: | 2004-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Bicarbonate Activation of Adenylyl Cyclase Via Promotion of Catalytic Active Site Closure and Metal Recruitment
Nat.Struct.Mol.Biol., 12, 2005
|
|
1QR6
 
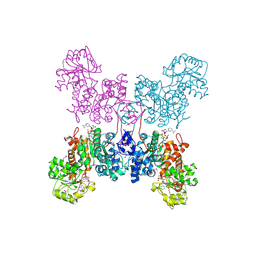 | | HUMAN MITOCHONDRIAL NAD(P)-DEPENDENT MALIC ENZYME | | Descriptor: | MALIC ENZYME 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Xu, Y, Bhargava, G, Wu, H, Loeber, G, Tong, L. | | Deposit date: | 1999-06-18 | | Release date: | 1999-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human mitochondrial NAD(P)(+)-dependent malic enzyme: a new class of oxidative decarboxylases.
Structure, 7, 1999
|
|
6CBY
 
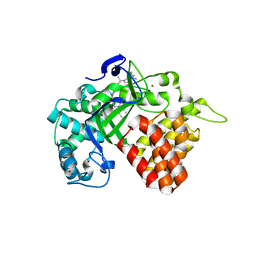 | | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF9975 | | Descriptor: | N-lysine methyltransferase SMYD2, ZINC ION, [3-(4-amino-6-methyl-1H-imidazo[4,5-c]pyridin-1-yl)-3-methylazetidin-1-yl][1-({1-[(1R)-cyclohept-2-en-1-yl]piperidin-4-yl}methyl)-1H-pyrrol-3-yl]methanone | | Authors: | ZENG, H, DONG, A, Hutchinson, A, Seitova, A, TATLOCK, J, KUMPF, R, OWEN, A, TAYLOR, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-05 | | Release date: | 2018-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Selective, Small-Molecule Co-Factor Binding Site Inhibition of a Su(var)3-9, Enhancer of Zeste, Trithorax Domain Containing Lysine Methyltransferase.
J.Med.Chem., 62, 2019
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4DZA
 
 | |
3EAE
 
 | | PWWP domain of human hepatoma-derived growth factor 2 (HDGF2) | | Descriptor: | Hepatoma-derived growth factor-related protein 2 | | Authors: | Amaya, M.F, Zeng, H, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
6JON
 
 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Primase | | Authors: | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6JOP
 
 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | Descriptor: | MAGNESIUM ION, Primase, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
4EA3
 
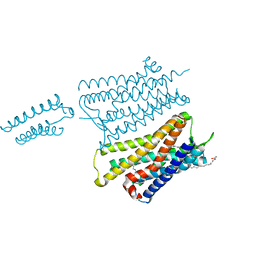 | | Structure of the N/OFQ Opioid Receptor in Complex with a Peptide Mimetic | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-benzyl-N-[3-(1'H,3H-spiro[2-benzofuran-1,4'-piperidin]-1'-yl)propyl]-D-prolinamide, ... | | Authors: | Thompson, A.A, Liu, W, Chun, E, Katritch, V, Wu, H, Vardy, E, Huang, X.P, Trapella, C, Guerrini, R, Calo, G, Roth, B.L, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic.
Nature, 485, 2012
|
|
2BBZ
 
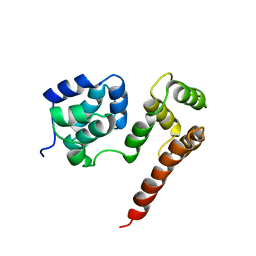 | | Crystal Structure of MC159 Reveals Molecular Mechanism of DISC Assembly and vFLIP Inhibition | | Descriptor: | Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Yang, J.K, Wang, L, Zheng, L, Wan, F, Ahmed, M, Lenardo, M.J, Wu, H. | | Deposit date: | 2005-10-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition.
Mol.Cell, 20, 2005
|
|
3DLM
 
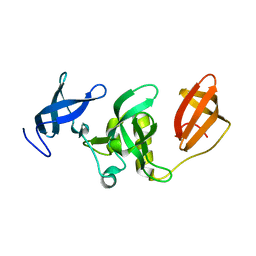 | | Crystal structure of Tudor domain of human Histone-lysine N-methyltransferase SETDB1 | | Descriptor: | Histone-lysine N-methyltransferase SETDB1 | | Authors: | Amaya, M.F, Dombrovski, L, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The crystal structure of Tudor domain of human Histone-lysine N-methyltransferase SETDB1.
To be Published
|
|
